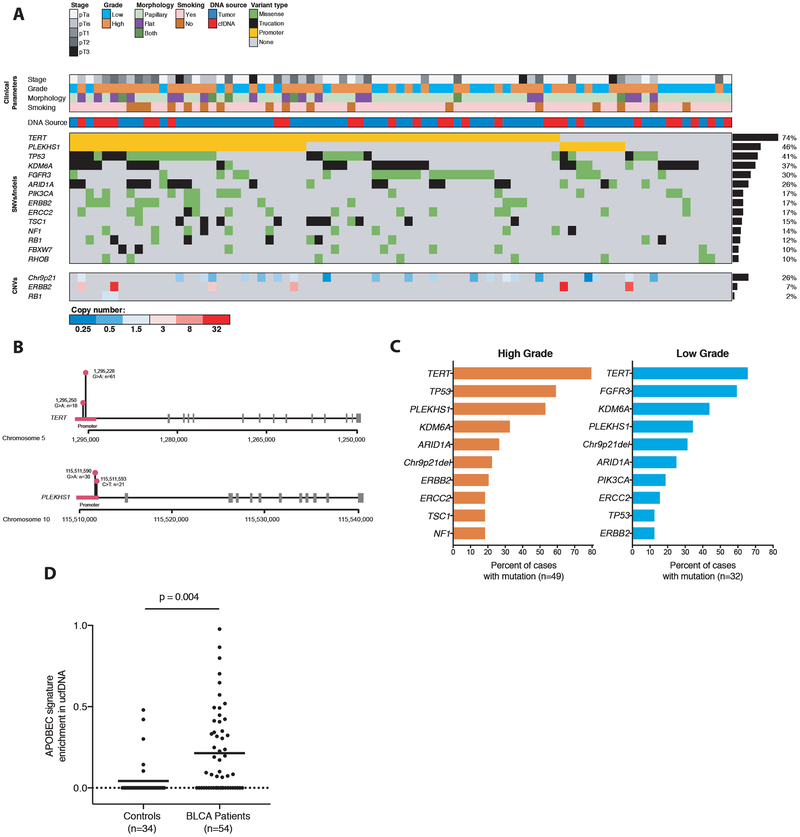
Figure 2: Genetic findings across bladder cancers profiled in study.
(A) Spectrum of genetic mutations and copy-number changes observed across 81 tumor and utDNA cases in this study, with clinicopathologic correlates. All tumor cases and all utDNA cases from patients with active cancer and at least one variant detected by genotyping were included in this analysis. All genes mutated in ≥ 10% of cases are shown, as well as all genes evaluated for copy-number variants. (B) Distribution of mutations in the TERT and PLEKHS1 promoters. (C) Comparison of mutations across high vs. low grade bladder cancers profiled in this study. (D) Enrichment of the APOBEC mutational signature in the cfDNA of patients with active bladder cancer versus healthy controls. P-values were calculated by multivariate regression controlling for total mutation count, median deduplicated sequencing depth, and the interaction between the two. APOBEC, apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like.