Figure 4.
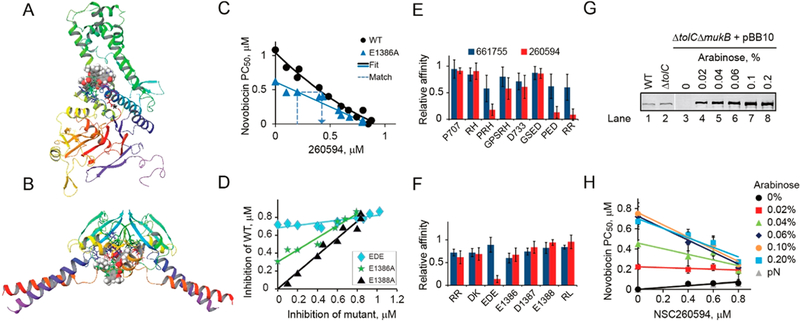
Binding sites of Michellamine B and 260594. (A, B) Predicted binding sites for Michellamine B and NSC260594 in the head (A) and hinge (B) domains. Compounds are shown in spheres; side chains are shown for residues in the binding sites. (C) Effect of NSC260594 on novobiocin susceptibility of cells with the wild type and a mutant MukB. Potentiation concentration PC50 of novobiocin was measured in the presence of the indicated amounts of the hit compound. The mutant and wild type data series were then matched by finding pairs of NSC260594 concentrations that produced the same novobiocin susceptibility in the two data sets and analyzed as illustrated in panel D. (D) NSC260594 inhibition curves for three representative mutants of MukB. The matching compound concentrations found as in panel C were plotted against each other and fit to a straight line. The slope of the fit line provides the upper bound on the ratio of the compound affinities to the mutant and wild type MukB. (E, F) Relative affinities of the hinge (E) and head (F) domain MukB mutants to the indicated hit compounds. (G) Immunoblotting analysis of MukB expression in BW25113 (WT), ETBW (ΔtolC), and OU142 (ΔtolC ΔmukB) cells harboring MukB-producing pBB10 plasmid. Lanes 1−3 and 4−10 contain extract form 0.1 and 0.004 OD of cells, respectively. MukB was detected using anti-MukB antibody.26 (H) Dose-dependent susceptibility of MukB overproducing OU142 cells harboring pBB10 plasmid to NSC260594, measured as the potentiation concentration of novobiocin in the presence of the indicated amounts of the compound and arabinose. pN, empty plasmid.
