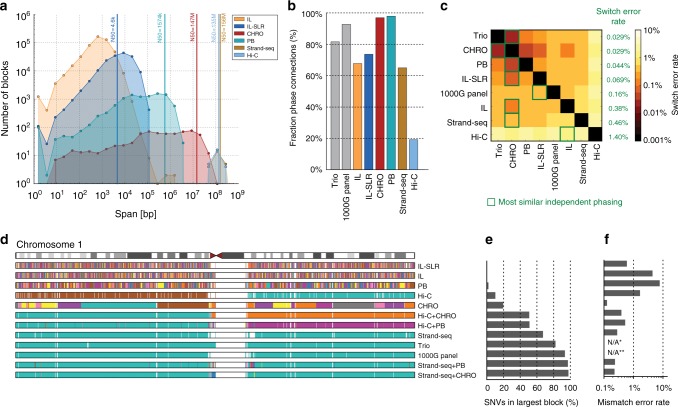
Fig. 1.
Characteristics of SNV-based haplotypes obtained from different data sources. a Distribution of phased block lengths for the YRI child NA19240. Note that Strand-seq haplotypes span whole chromosomes and therefore one block per chromosome is shown. Vertical bars highlight N50 haplotype length: the minimum length haplotype block at which at least half of the phased bases are contained. For Illumina (IL) paired-end data, phased blocks cover <50% of the genome and hence the N50 cannot be computed. b Fraction of phase connection, i.e., pairs of consecutive heterozygous variants provided by each technology (averaged over all proband samples). c Pairwise comparisons of different phasings; colors encode switch error rates (averaged over all proband samples). For each row, a green box indicates the phasing of an independent technology with best agreement, with corresponding switch error rates given in green. d Each phased block is shown in a different color. The largest block is shown in cyan, i.e., all cyan regions belong to one block, even though interspaced by white areas (genomic regions where no variants are phased) or disconnected small blocks (different colors). e Fraction of heterozygous SNVs in the largest block shown in d. f Mismatch error rate of largest block compared to trio-based phasing, averaged over all chromosomes of all proband genomes (i.e., the empirical probability that any two heterozygous variants on a chromosome are phased correctly with respect to each other, in contrast to the switch error rate, which relays the probability that any two adjacent heterozygous variants are phased correctly). (*) Not available because trio phasing is used as reference for comparisons. (**) Not shown as population-based phasing does not output block boundaries; refer to Supplementary Material for an illustration of errors in population-based phasing