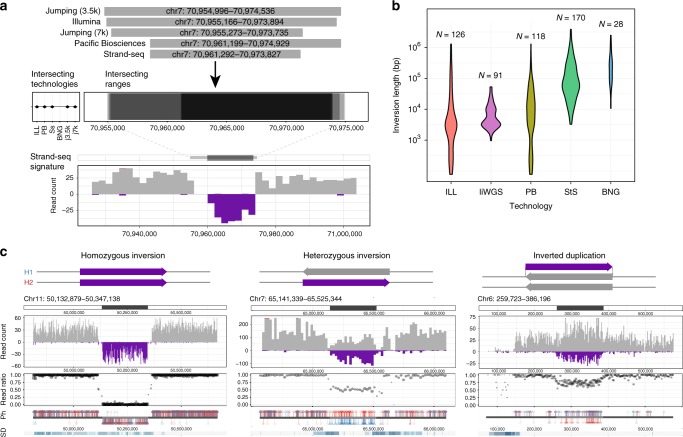
Fig. 3.
Characterization of simple and complex inversions. a Integration of inversions across platforms based on reciprocal overlap. Shown is an example of five orthogonal platforms intersecting at a homozygous inversion, with breakpoint ranges and supporting Strand-seq signature illustrated in bottom panels. b Size distribution of inversions included in the unified inversion list, subdivided by technology, with the total inversions (N) contributed by each listed. c Classification of Strand-seq inversions based on orthogonal phase support. Illustrative examples of simple (homozygous and heterozygous) and complex (inverted duplication) events are shown. Strand-seq inversions were identified based on read directionality (read count; reference reads in gray, inverted reads in purple), the relative ratio of reference to inverted reads within the locus (read ratio), and the haplotype structure of the inversion, with phased read data considered in terms of directionality (Ph; H1 alleles in red, H2 alleles in blue; alleles from reference reads are displayed above the ideogram and alleles from inverted reads are displayed below). ILL Illumina. liWGS long-insert whole-genome sequencing libraries. PB Pacific Biosciences. StS Strand-seq. BNG Bionano Genomics. SD segmental duplication. Ph phase data