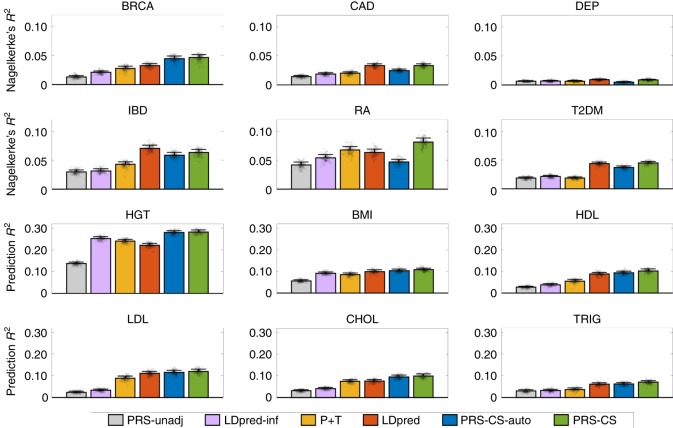
Fig. 2.
Prediction accuracy of six polygenic prediction methods in the Partners HealthCare Biobank. Posterior effect sizes of single nucleotide polymorphisms (SNPs) were trained with large-scale genome-wide association summary statistics, using the 1000 Genomes Project European sample as an external linkage disequilibrium (LD) reference panel. Polygenic scores were applied to predict six curated common complex diseases—breast cancer (BRCA), coronary artery disease (CAD), depression (DEP), inflammatory bowel disease (IBD), rheumatoid arthritis (RA), and type 2 diabetes mellitus (T2DM), and six quantitative traits—height (HGT), body mass index (BMI), high-density lipoproteins (HDL), low-density lipoproteins (LDL), cholesterol (CHOL), and triglycerides (TRIG). The Partners HealthCare Biobank sample for each disease and quantitative phenotype was repeatedly and randomly split into a validation set comprising 1/3 of the data and a testing set comprising 2/3 of the data. Tuning parameters (P-value threshold in P+T, fraction of causal SNPs in LDpred, and global shrinkage parameter in PRS-CS) were selected in the validation data set, and the predictive performance was assessed in the testing set. For disease (case–control) phenotypes and quantitive traits, prediction accuracy was measured by the Nagelkerke’s R2 and R2, respectively, averaged across 100 random splits. The error bar indicates the standard deviation of prediction accuracy across 100 random splits. Prediction accuracy for each random split is overlaid on the bar plot (black circles)