Figure 2.
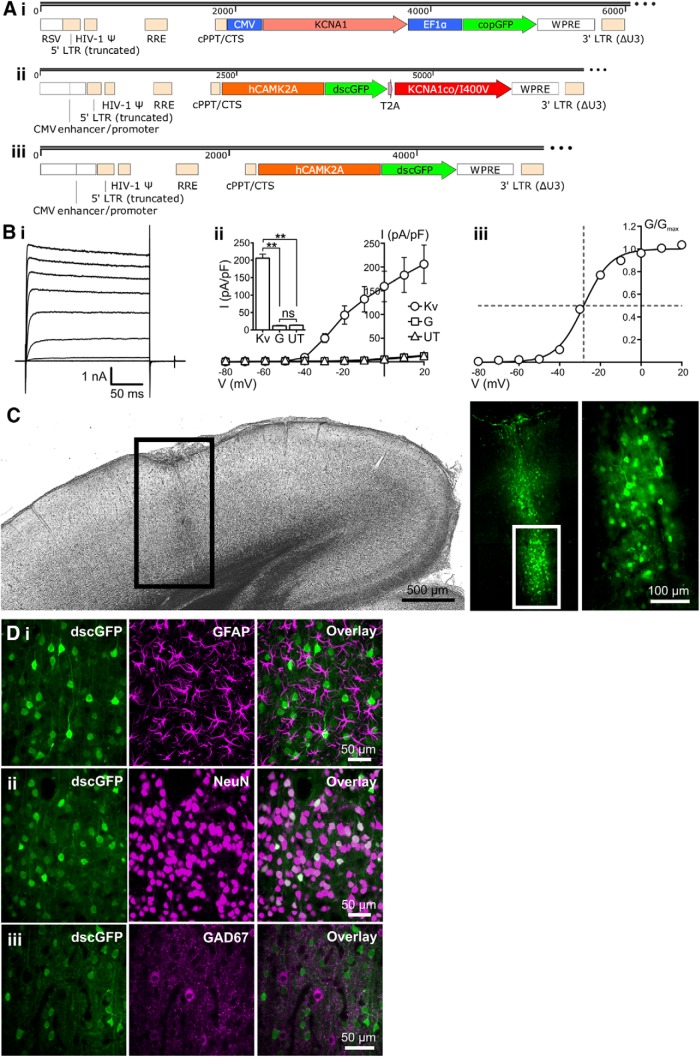
Design and characterization of an EKC gene therapy optimized for clinical translation. A, Transfer plasmid maps for the Lenti-CMV-KCNA1 pilot vector (i), the optimized Lenti-CaMKII-EKC vector (ii), and its Lenti-CaMKII-GFP control (iii). RSV, Rous sarcoma virus promoter; LTR, long terminal repeat; HIV-1 Ψ, HIV-1 packaging signal; RRE, Rev response element; cPPT/CTS, central polypurine tract and central termination sequence; EF1α, elongation factor 1 α promoter; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element. B, Heterologous expression of functional Kv1.1 channels from the optimized Lenti-CaMKII-EKC transfer plasmid. i, Representative current–time trace from a Neuro-2a cell transfected with the Lenti-CaMKII-EKC transfer plasmid. ii, Plot of mean current density against voltage for cells transfected with the Lenti-CaMKII-EKC transfer plasmid (Kv; n = 13), cells transfected with the Lenti-CaMKII-GFP control plasmid (G; n = 8), and untransfected controls (UT; n = 10). Inset, Histogram showing differences in current density among the three groups during the voltage step to +20 mV (Kv vs UT, p = 0.0013; Kv vs G, p = 0.0012; UT vs G, p = 0.82; ns = not significant; Welch's one-way ANOVA with Games–Howell post hoc tests). iii, Plot of mean normalized conductance against voltage for cells transfected with the Lenti-CaMKII-EKC transfer plasmid. Data are fit with a single Boltzmann function. The V0.5 value of −28.2 mV is similar to values obtained from HEK293 cells transfected with CMV-driven, wild-type KCNA1 (−32.8 ± 0.9 mV; Tomlinson et al., 2013). All error bars represent the SEM. C, Bright-field and fluorescence images of a brain slice from a rat injected in the left visual cortex with 1.25 μl (∼3.0 × 106 IU) of the Lenti-CaMKII-EKC vector. The pattern of transduction is similar to that observed with the Lenti-CMV-KCNA1 vector. D, Immunohistochemical assessment of the cell type specificity of EKC expression. i, There was no overlap between transduced neurons expressing dscGFP and astrocytes stained for GFAP. ii, There was 100% overlap between dscGFP+ cells and neurons stained for NeuN. iii, Minimal overlap was observed between dscGFP+ cells and inhibitory interneurons stained for GAD67. **p < 0.002.