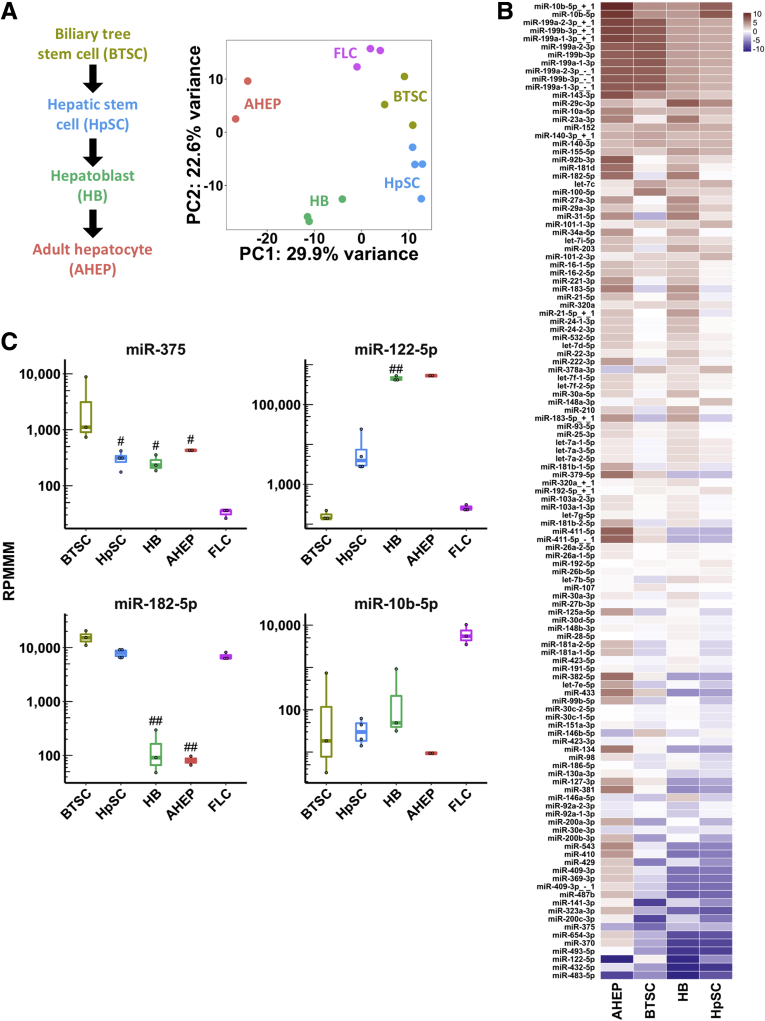
Figure 2.
miR-375 is dramatically suppressed in an FLC patient-derived xenograft model relative to every lineage stage of the liver. (A) Maturational trajectory depicts the lineage stages within the liver (left). Principal component analysis of small RNA-seq data from a FLC PDX model and 4 maturational lineage stages of the liver (right). (B) Heatmap showing miRNA expression in the FLC PDX model. Color intensity shows log2 (fold change) relative to 4 maturational lineage stages of the liver. miRNAs are included in the heatmap if they had an average expression >1000 reads per million mapped to miRNAs (RPMMM) in any cell type and P < .05 in any cell type compared with FLC. (C) Expression of 2 candidate tumor-suppressor miRNAs (miR-375 and miR-122-5p) and 2 candidate oncomiRs (miR-182-5p and miR-10b-5p) from small RNA-seq in 4 maturational lineage stages of the liver and the FLC PDX model. #P < .05, ##P < .01 (2-tailed Student t test; P ≥ .05, Mann–Whitney U test). PC, principal component.