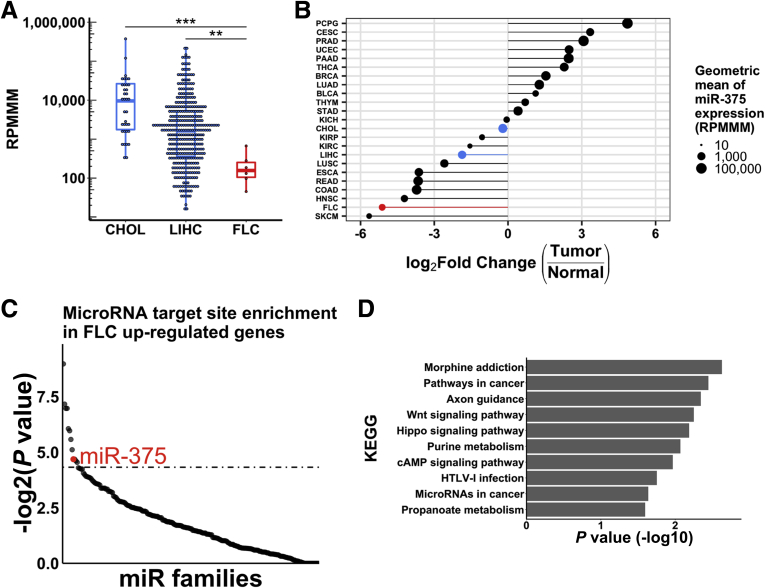
Figure 4.
miR-375 is a candidate master regulator of cancer pathways in FLC. (A) Expression of miR-375 in CHOL (n = 36), LIHC (n = 366), and FLC (n = 6) in TCGA smRNA-seq data. (B) Log2 (fold change) of miR-375 expression in different tumor types (n = 23) within TCGA. The size of each circle represents the geometric mean of miR-375 expression in each tumor type. Each tumor type is ranked on the y-axis by the log2 (fold change) of the geometric mean of tumor expression over non–tumor expression of miR-375. Geometric means were used instead of arithmetic means to provide robustness to outliers. Highlighted are FLC (red) as well as CHOL and LIHC (blue). (C) Ranked –log2 (P value) of miRhub Monte Carlo simulation. miRNAs were examined for target site enrichment in genes up-regulated in FLC. Dotted line represents P = .05. (D) KEGG enrichment analysis of up-regulated miR-375 target genes in FLC. **P < .01, ***P < .001 (Mann–Whitney U test, 2-sided). BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal papillary cell carcinoma; KIRP, kidney renal clear cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma.