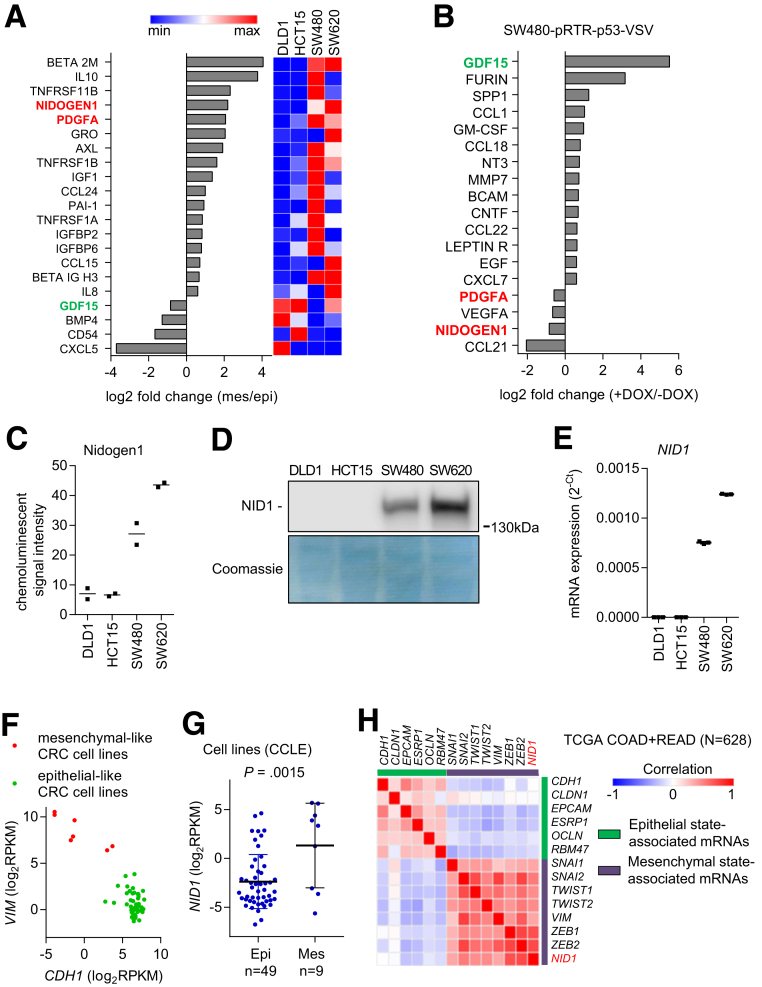
Figure 4.
Identification of EMT regulators within CM of CRC cell lines. (A) Cytokine array analyses of CM from epithelial (DLD1 and HCT15) and mesenchymal (SW480 and SW620) CRC cell lines. Secreted factors that showed at least 1.5-fold difference between epithelial-like and mesenchymal-like CRC cell lines are shown. (B) Cytokine array analyses of CM from SW480-pRTR-p53-VSV cells. Secreted factors that showed at least 1.5-fold difference between SW480-pRTR-p53-VSV cells treated with vehicle (p53 off) or DOX (p53 on) are shown. (C) NID1 levels in CM of indicated cell lines assessed by densitometric quantification of NID1-specific signals from cytokine array analyses. (D) Western blot analysis of NID1 protein in concentrated CM from indicated cell lines. Coomassie Blue staining was used as a loading control. (E) qPCR analyses of NID1 expression in indicated cell lines. (F) Classification of colorectal cancer cell lines as epithelial- or mesenchymal-like based on the distribution of vimentin (VIM) and E-cadherin (CDH1) mRNA expression. Data are from the Cancer Cell Line Encyclopedia database (CCLE). (G) NID1 expression in epithelial- and mesenchymal-like CRC cell lines represented in the CCLE database. Individual data points and means ± SD are provided. (H) Correlation of NID1 expression with epithelial- and mesenchymal-state–associated mRNAs in primary CRC tumors. Expression data are from the TCGA collection of human colorectal adenocarcinomas (COAD + READ; N = 628). (C) Individual values and the means (n = 2 technical replicates/1 biological sample), and (E) means ± SD (n = 3 biological replicates) are provided. Epi, epithelial-like CRC cell lines; Mes, mesenchymal-like CRC cell lines.