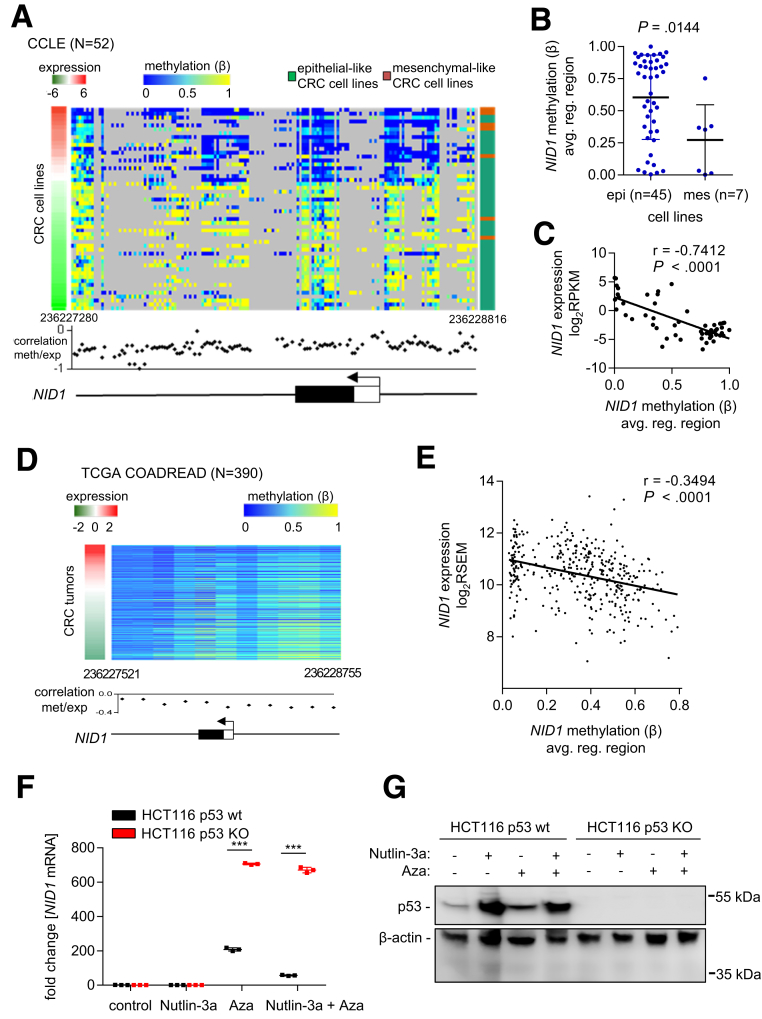
Figure 6.
Expression of NID1 is suppressed by DNA methylation. (A) DNA methylation and expression of NID1, as well as epithelial/mesenchymal status of 52 CRC cell lines (data from the Cancer Cell Line Encyclopedia). Left: red/green colored bar: NID1 expression; middle: blue/yellow heatmap: DNA methylation (β value) for each CG site in the NID1 regulatory genomic region; right: green/orange bar: epithelial/mesenchymal status of cell lines. The graph below the heatmap shows the correlation of each CG site in the NID1 regulatory genomic region with NID1 expression. hg19 genomic coordinates and the NID1 gene structure with indicated first exon (white box: untranslated region; black box: protein encoding region) and transcription start site (arrow) is shown at the bottom. (B) Average DNA methylation of all CG sites in the NID1 regulatory genomic region in epithelial-like and mesenchymal-like CRC cell lines (data from the Cancer Cell Line Encyclopedia; Student t test). Individual data points and means ± SD are provided. (C) Correlation between average DNA methylation of all CG sites in the NID1 regulatory gene region and NID1 expression in 52 CRC cell lines (data from the Cancer Cell Line Encyclopedia). Spearman correlation coefficient is provided. (D) DNA methylation and expression of NID1 in 390 primary colorectal tumors (data from the TCGA COAD + READ cohort). Left: red/green colored bar: NID1 expression; middle: blue/yellow heatmap: DNA methylation (β value) for each CG site in the NID1 regulatory genomic region. The graph bellow the heatmap shows the correlation of each CG site in the NID1 regulatory genomic region with NID1 expression. hg19 genomic coordinates and the NID1 gene structure with indicated first exon (white box: untranslated region; black box: protein encoding region) and transcription start site (arrow) is shown at the bottom. (E) Correlation between average DNA methylation of all CG sites in the NID1 regulatory genomic region and NID1 expression in primary colorectal tumors (data from the TCGA COAD + READ cohort). Spearman correlation coefficient is provided. (F) NID1 mRNA expression in HCT116 p53 wt and p53 knockout cells. Cells were treated with the DNA methyltransferase inhibitor 5-aza-2’-deoxycytidine for 96 hours and with Nutlin-3a for the last 48 hours. Significance was determined using 1-way analysis of variance with the Tukey multiple comparison post-test; ***P < .001. (G) Western blot analysis of p53 protein in HCT116 p53 wt and p53 knockout cells treated as described in panel F.