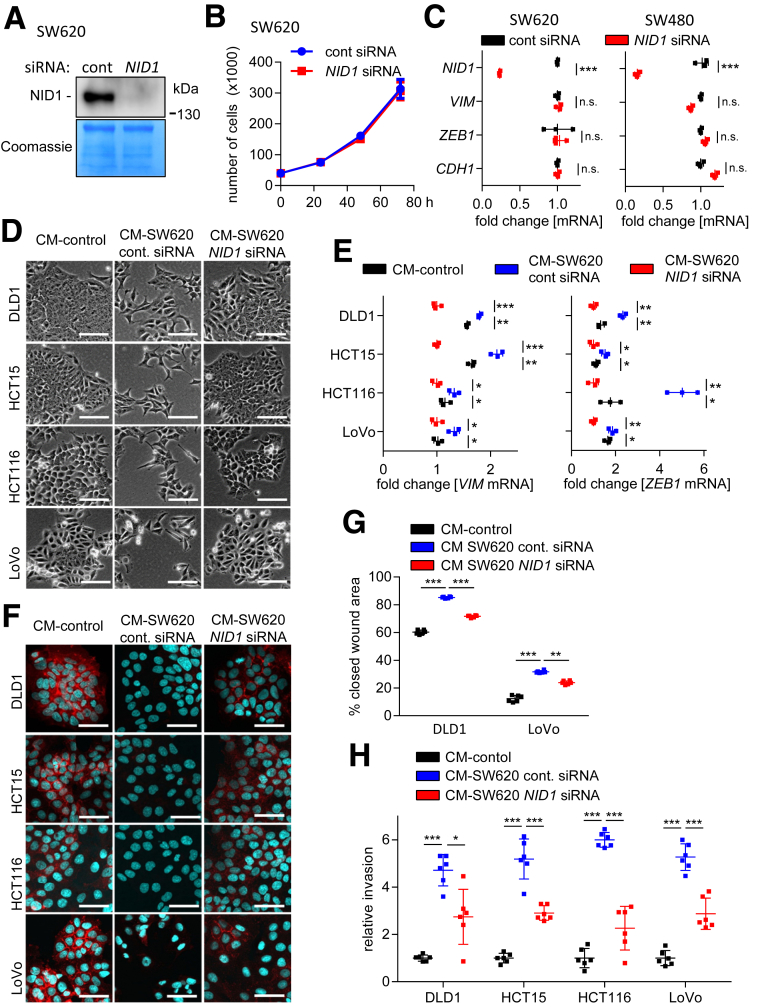
Figure 7.
Requirement of NID1 for CM-induced EMT, migration, and invasion. (A) Western blot analysis of NID1 protein in concentrated CM from SW620 cells transfected with control or NID1-specific siRNAs for 72 hours. Coomassie Blue staining was used as loading control. (B) Cell number of SW620 cells transfected with control or NID1-specific siRNAs at the indicated time points. (C) qPCR analysis of NID1 and EMT-related mRNAs ZEB1, VIM, and CDH1 in SW480 and SW620 cells transfected with control or NID1-specific siRNAs for 72 hours. (D) Phase-contrast images showing cell morphology of DLD1, HCT15, HCT116, and LoVo cells treated for 96 hours with CM from SW620 cells 72 hours after transfection with control or NID1-specific siRNAs. (E) qPCR analyses of VIM and ZEB1 in DLD1, HCT15, HCT116, and LoVo cells treated for 96 hours with CM collected from SW620 cells 72 hours after transfection with control or NID1 siRNA. (F) Immunofluorescence detections of E-cadherin in DLD1, HCT15, HCT116, and LoVo cells treated for 96 hours with CM collected from SW620 cells 72 hours after transfection with control or NID1 siRNA. Nuclear DNA was detected by staining with 4′,6-diamidino-2-phenylindole. Scale bar: 50 μm. (G) Wound healing assay of DLD1 and LoVo cells treated with indicated CMs for 30 hours. The width of scratches in 2 independent wells was analyzed for each state. Results represent the average (%) of wound closure. (H) Invasion assay in modified Boyden chambers: DLD1, HCT15, HCT116, and LoVo cells were seeded on Matrigel-coated filters with indicated CMs used as a chemoattractant in the lower well. After 48 hours, cells that invaded through the Matrigel were counted after 4′,6-diamidino-2-phenylindole staining. Means ± SD of (B, C, and E) n = 3 biological replicates, and (G and H) n = 2 biological replicates (each n = 3 technical replicates) are provided. Significance was determined using 1-way analysis of variance with the Tukey multiple comparison post-test; *P < .05; **P < .01; ***P < .001. cont, control.