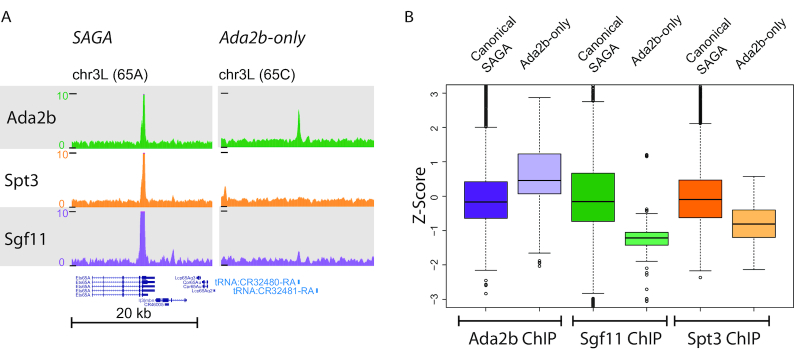
Figure 1.
A subset of Ada2b binding sites do not correspond to canonical SAGA sites. (A) Genome browser screenshot shows genes bound by canonical SAGA (Ada2b, Spt3, Sgf11), and genes bound exclusively by Ada2b (note the absence of Spt3 and Sgf11 binding). (B) Box plots of the Z-scores of the canonical SAGA peaks and Ada2b-only peaks for subunits in the HAT, SPT and DUB modules of SAGA. Each boxplot shows the Z-score for either canonical SAGA or Ada2b-only loci. Canonical SAGA medians are centered around zero, indicating that their distribution is similar comparable to the Z-score distribution of the peak enrichments observed in the reference peak dataset. The distribution of the Ada2b signal for the Ada2b-only peaks is positively skewed, indicating that Ada2b is enriched at these sites. In contrast, the Sgf11 and Spt3 distribution was skewed to a value ≤-1, indicating that these factors are not enriched at the Ada2b-only peaks.