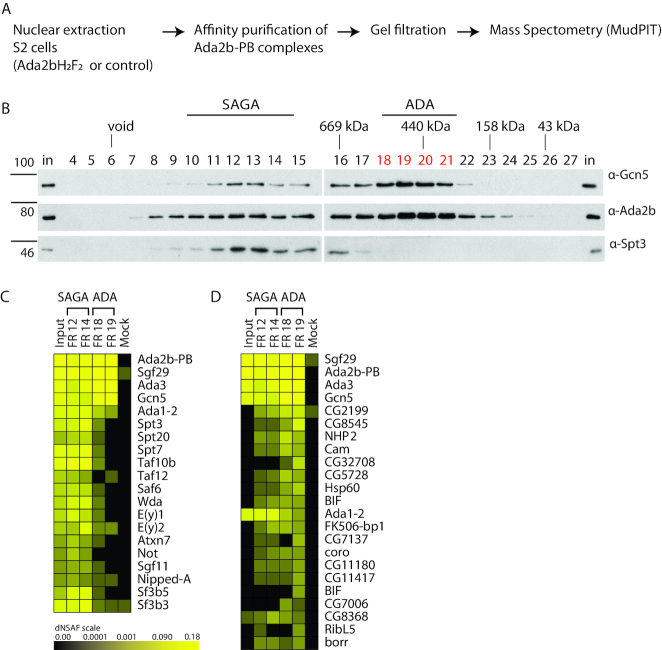
Figure 4.
Affinity purification of the small ADA complex from S2 cell nuclear extract for MudPIT analysis. (A) Workflow. Ada2b-PB complexes were affinity purified from nuclear extract of S2 cells expressing Ada2b-PBH2F2 and separated by gel filtration. Mock affinity purifications were performed on nuclear extract from cells that do not express a Flag-tagged protein. Select samples were subjected to MudPIT analysis. (B), Flag affinity-purified Ada2b-PB complexes were subjected to gel filtration by Superose 6, and samples of each fraction were probed for Gcn5, Ada2b and Spt3 by Western blot. As in fractionated crude nuclear extract, two Ada2b peaks were observed. The SAGA complex eluted in fractions 10–14. The small ADA complex eluted in fractions 18–21. Unfractionated affinity-purified Ada2b complexes (‘input’), SAGA fractions 12 and 14, ADA fractions 18 and 19, and mock purification samples were subjected to MudPIT. (C and D) Heatmaps show relative protein abundance expressed as dNSAF (distributed normalized spectral abundance factor) with the brightest yellow indicating high abundance and decreasing intensity reflecting progressively lower abundance. The abundance of various SAGA subunits in the affinity-purified FLAG elution input for gel filtration, gel filtration fractions 12 and 14, gel filtration fractions 18 and 19, and averaged mock samples are presented as columns. Individual proteins are as indicated in each row. (C) The abundance of SAGA subunits in the high molecular weight fractions compared to those in the smaller ADA complex, along with input and mock controls. Subunit composition of the SAGA complex fractions show that Ada2b purification pulls down known SAGA subunits, indicating that the passage over the gel filtration column does not affects the detection of the subunits. (D) Non- SAGA subunits that uniquely co-purify with the ADA complex. The dNSAF values for fraction 18 and 19 were averaged and sorted in decreasing order. Proteins also enriched in the mock sample were omitted. No additional proteins co-purified uniquely with Ada2b in the ADA complex. Only Sgf29, Gcn5 and Ada3 are strongly enriched in this complex. Other proteins are either 1) not specific to the ADA complex and copurify with SAGA 2) have low dNSAF values in the ADA complex and/or 3) are common contaminants.