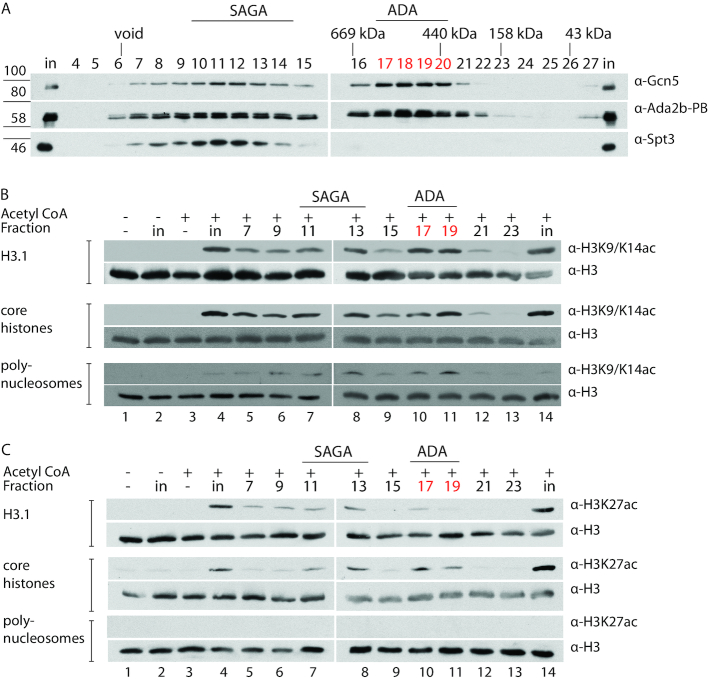
Figure 5.
The Drosophila ADA complex has histone acetyltransferase activity in vitro. Ada2b-PB complexes were affinity purified overnight from nuclear extract of S2 cells expressing Ada2b-PBH2F2 and separated by gel filtration. (A) The gel filtration profile of Gcn5, Ada2b and Spt3. The SAGA complex eluted in fractions 10–14, and the ADA complex in fractions 17–20. B, H3K9/14 HAT assays using fractions from A that correspond to the Gcn5-containing SAGA and ADA complexes. Column fractions of affinity-purified Ada2b-PB were tested for HAT activity on recombinant human histone 3.1 (top two panels), HeLa cell core histones (middle two panels), and HeLa cell polynucleosomes (bottom two panels). Negative controls included histone substrate only, to control for background histone acetylation (lane 1), input without Acetyl-CoA to control for spurious HAT activity (lane 2), and Acetyl-CoA without HAT-containing extract to control for nonspecific acetylation due to excess substrate and cofactor (lane 3). Lane 4 includes a positive control in which the HAT assay was performed with the gel filtration input prior to size-separation of the Ada2b-PB-containing complexes. For all histone substrates, peaks of H3K9/H3K14 acetyltransferase activity were observed in fraction 13, coinciding with the SAGA complex, and fractions 17 and 19, coinciding with the ADA complex. (C) H3K27 HAT assays using Gcn5-containing SAGA and ADA complexes. Column fractions of affinity-purified Ada2b-PB were tested for HAT activity with the substrates and controls described in (B) above. No H3K27 acetylation was observed for the H3.1 and core histone controls (lanes 1–3), indicating that the antibody is specific, and does not detect H3K27ac that is naturally present on acid-extracted core histones. H3K27 acetylation is highest in the SAGA and ADA-containing fractions, correlating with the amount of Gcn5. In contrast, neither the ADA complex (lanes 10 and 11) nor SAGA complex (lane 7 and 8) acetylate lysine 27 on polynucleosomes.