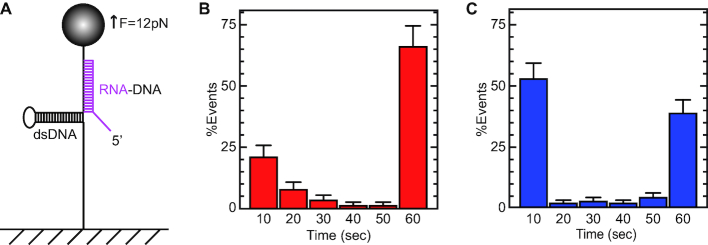
Figure 5.
Experimental protocol for determining the processivity of Ded1. (A) An RNA oligonucleotide (magenta) was used that could form a 30 bp duplex with the DNA and that had a 5′, 15-nucleotide-long, single-stranded region. The RNA hybridized in the forked region of the DNA hairpin, and consequently the hairpin could only partially reform. An Fhold of 12 pN was used to offset the refolding force of the hairpin that would otherwise displace the hybridized RNA (equivalent to step C, Figure 1A). The Fhold was maintained for a Thold of 60 s. The force was then reduced to zero, and the oligonucleotide was fully displaced for the next round of experiments. (B) The distribution of the Toff values of the oligonucleotide in the absence of protein. Each of the histogram bars represents the mean and standard deviations of oligonucleotides displaced from the DNA at the indicated times that was normalized for the total observed events. The distribution was not uniform; this indicated that the long-lived species were stable during the hold time of 60 seconds and represented fully hybridized duplexes. The shorter-lived species (10–30 s) represented partially hybridized oligonucleotides that were eventually displaced by the refolding DNA hairpin. (C) The displacement profile of the hybridized oligonucleotide in the presence of 100 nM Ded1 and 1 mM ATP. Ded1 facilitated the displacement of the partially hybridized oligonucleotides, which resulted in a single large peak at 10 s. In contrast, Ded1 was not able to displace the fully hybridized oligonucleotide during the duration of the Thold. The mean number of events for each time interval normalized for the total number of observed events is shown with the standard deviations. The lower error bars were deleted for clarity.