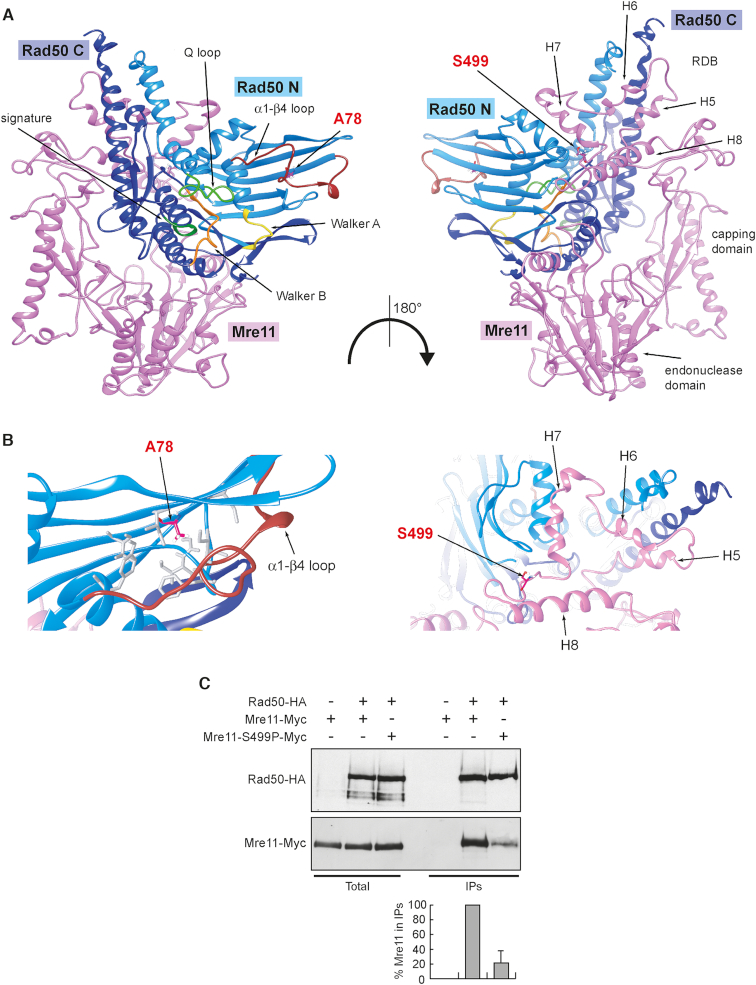
Figure 5.
The mre11-S499P mutation is localized at the Mre11–Rad50 interface and reduces Mre11–Rad50 interaction. (A) Structural prediction of S. cerevisiae Mre11–Rad50 heterodimer, obtained as previously described (38), showing the localization of S499 residue in the Rad50-binding domain (RBD) of Mre11 and of A78 residue in the N-terminal of Rad50. Pink, Mre11. Light blue, N-terminal lobe of Rad50 (Rad50 N). Dark blue, C-terminal lobe of Rad50 (Rad50 C). (B) Detailed view of the molecular surroundings of the residues affected by the mutations. The hydrophobic residues surrounding A78 in Rad50 are shown as gray sticks. (C) Mre11–Rad50 interaction. Protein extracts prepared from exponentially growing cells were analyzed by western blotting with anti-HA and anti-Myc antibodies either directly (Total) or after immunoprecipitation (IPs) with anti-HA antibody. Graph represents the amount of Mre11-S499P-Myc relative to Mre11-Myc that was set up to 100%. Plotted values are the mean values with error bars denoting S.D. (n = 3). An immunoblot from one of these experiments is shown.