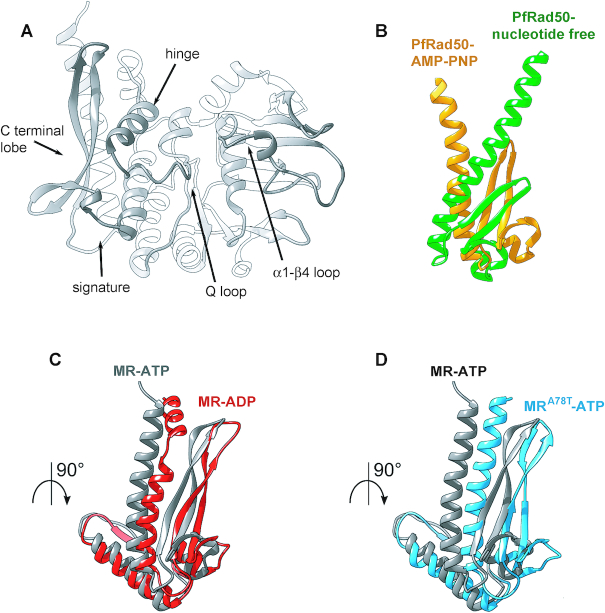
Figure 8.
The rad50-A78T mutation induces similar conformational changes in Rad50 as ATP hydrolysis. (A) Cartoon of the centroid structures of one Rad50 subunit (chains C and D) from the absolute minimum corresponding clusters from MR-ATP showing the main features involved in the conformational change upon ADP versus ATP binding. (B) View of the C-terminal lobes of the structure of P. furiosus Rad50 bound to AMP-PNP (PDB:3qku; orange) superimposed to nucleotide-free Rad50 (PDB:3qks; green). (C, D) View of the C-terminal lobes of centroid structures of the energetic absolute minimum of Rad50 subunit from MR-ATP MD simulations (gray) superimposed by structural alignment to the N-terminal region of MR-ADP (C) and MRA78T-ATP (D) energetically favourite Rad50 structures.