Figure 4.
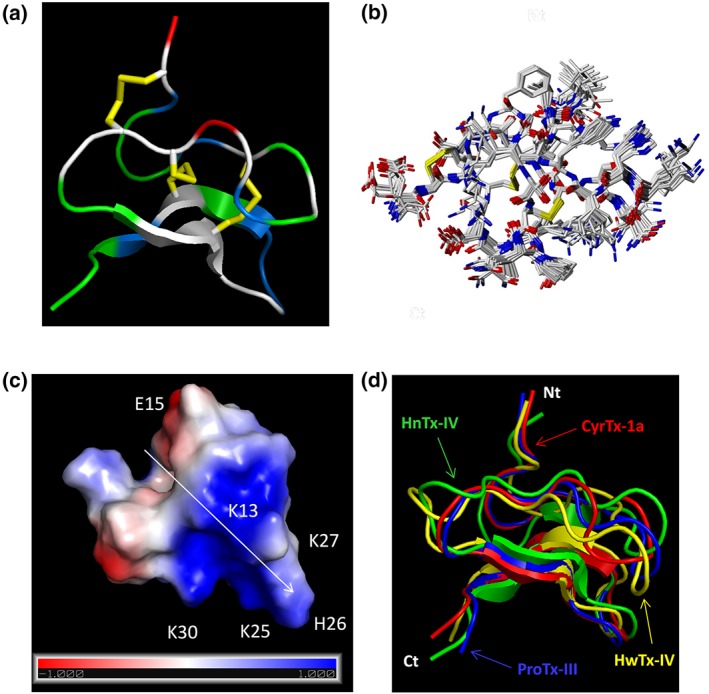
Representation of CyrTx‐1a identified by PyMOL. (a) Representation of the backbone peptide folding of CyrTx‐1a determined by 1H 2D NMR method. The structure topology is composed of double stranded antiparallel β sheet. The three disulfide bonds are C2–C17, C9–C22, and C16–C29 (in yellow). Hydrophobic residues are coloured in green, and basic and acidic residues are coloured in blue and red, respectively. The other polar residues are coloured in white. (b) Superposition of 20 structures derived from a 6‐ns restrained MD simulation (all heavy atoms are shown). All backbone atoms of residues two to 31 were used for fitting. Structures were sampled in 300 ps intervals and energy minimized. The rmsd over all backbone atoms (including residues one to 33) is 0.465 Å with an SD of 0.157 Å. Considering all heavy atoms, the rmsd is 1.072 Ǻ with an SD of 0.285 Ǻ (PDB: 6GFT). (c) Electrostatic charged surface representation of CyrTx‐1a. The molecule is rendered as a surface coloured according to the electrostatic potential. As indicated in the coloured legend, an excess of negative and positive charges near the surface are represented in red (−1,000) and blue (1,000), respectively, while fairly neutral potentials are represented in white. The entire structure has a clear dipole potential with E1 and E15 forming a negative zone while K3, K7, K13, K25, H26, K27, and K30 form a positive zone. (d) Superposition of backbone peptide folding of CyrTx‐1a and three other toxins of the NaSpTx family 1 previously described to possess analgesic effects (PDB entries of HnTx‐IV: 1NIY, ProTx‐III: 2MXM, and HwTx‐IV: 1MB6)
