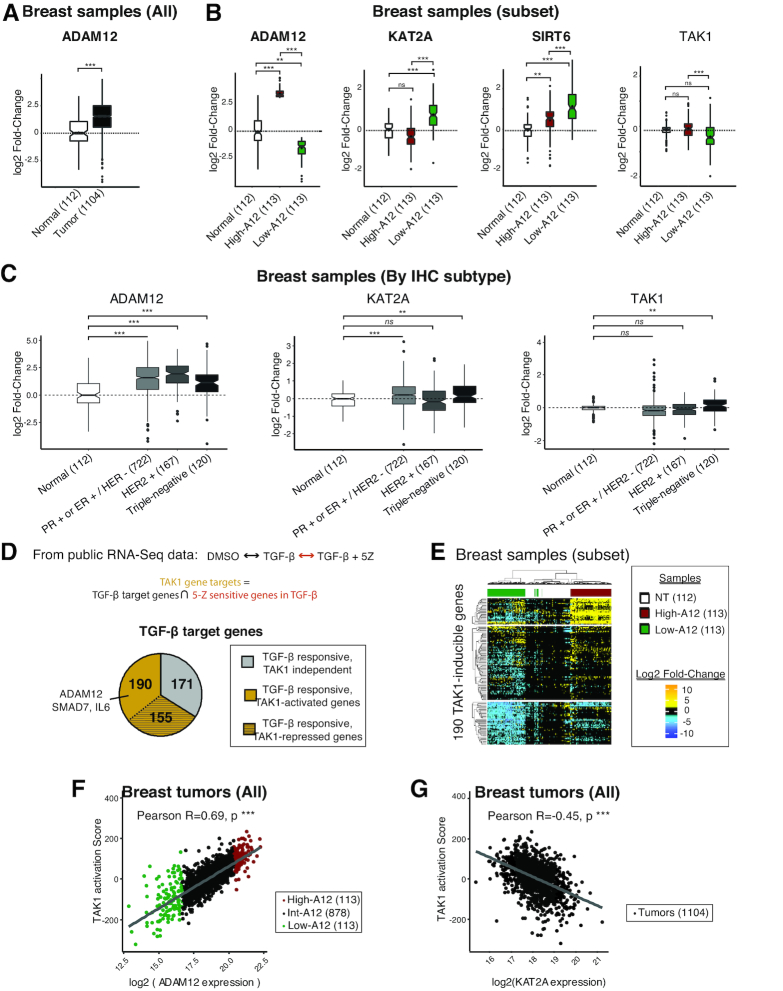
Figure 7.
TCGA analysis revealed co-expression of ADAM12 with KAT2A, TAK1 and TAK1-inducible genes signature in breast tumors. (A) ADAM12 expression in breast tumors and normal breast tissue from the TCGA database. The boxplots contain 50% of the values, with a notch at the median value, and a diamond at the average value. The whiskers depict the first and last quartiles, and outliers appear as black dots. (B) ADAM12, KAT2A, and TAK1 expression in normal breast tissues and a subset of breast cancer samples from the TCGA database, selected based on their low (first decile, green) or high (ninth decile, red) ADAM12 expression. (C) ADAM12, KAT2A, and TAK1 expression in normal breast tissues and breast cancer samples from the TCGA database stratified by subtype. (D) Definition of the TAK1 signature, using transcriptomic data for primary cells treated with TGF-β with or without the TAK1 inhibitor 5Z. The TAK1 signature contains the genes that are upregulated by TGF-β addition, but not upregulated in the TGF-β+5Z condition. (E) Hierarchical clustering with euclidean distance metric and Ward's linkage method of highest and lowest ADAM12 expressing tumors samples, and normal breast tissue samples. The unsupervised clustering of breast tumors according to the TAK1 signature almost perfectly segregates tumors according to ADAM12 expression. (F) TAK1 activation score (calculated from the expression of genes in the TAK1 signature) correlates positively with ADAM12 expression in 1104 breast tumors. The high-ADAM12 samples are shown in red and low-ADAM12 in green. The statistical analysis was performed with a two-way ANOVA test, followed by a Tukey HSD test (*** denotes P < 0.001). (G) TAK1-activation score negatively correlates with KAT2A expression in 1104 breast tumors. Statistics as in panel E.