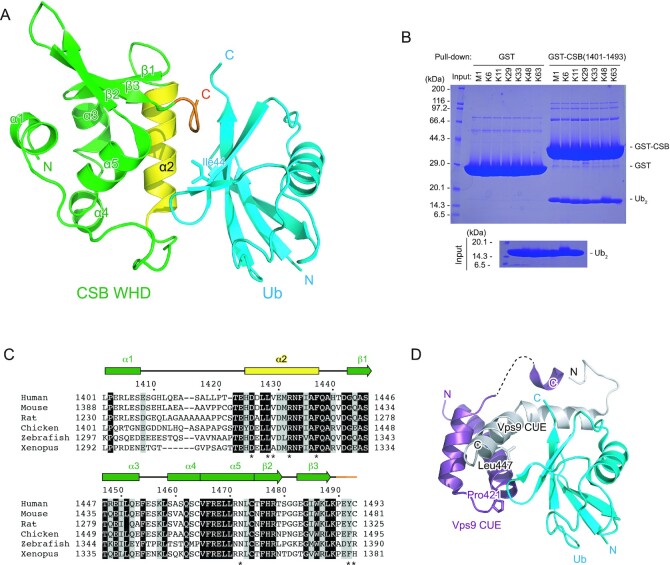
Figure 3.
Structure of the complex between CSB WHD and Ub. (A) Crystal structure of one CSB WHD in complex with Ub. The coloring scheme is the same as that in Figure 2. (B) Pull-down assays between CSB WHD (1401–1493) and M1-, K6-, K11-, K29-, K33-, K48- or K63-Ub2. The bound Ub2 species were analyzed by SDS-PAGE with Coomassie brilliant blue staining. (C) Sequence alignment of CSB WHD from human (NP_000115.1), mouse (NP_001074690.1), rat (NP_001100766.1), chicken (XP_421656.2), zebrafish (XP_688972.2) and Xenopus (NP_001016056.1). Multiple sequence alignment was performed using the program ClustalW (53). Fully conserved residues are colored white with black backgrounds, whereas residues with similar properties (scoring > 0.5 in the Gonnet matrix (54)) are marked with grey backgrounds. Asterisks represent the residues whose side chains interact with Ub. (D) Structure of the complex between Ub and the Vps9 CUE dimer. One Vps9 CUE molecule in the dimer is colored white, whereas the other molecule is colored purple. The side chains of Pro421 and Leu447 in Vps9 CUE are shown as sticks.