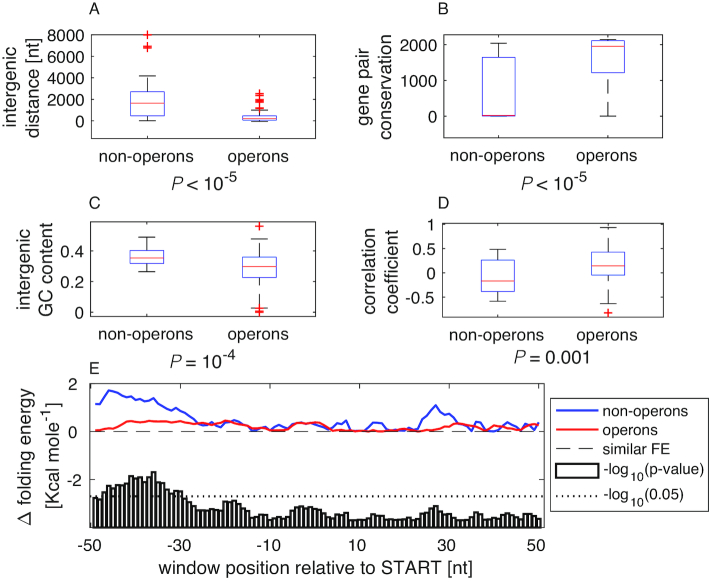
Figure 2.
Operon sequence features. Distribution of (A) intergenic distances, (B) gene pair conservation, (C) intergenic GC content, (D) RNA structure similarity (Pearson’ correlation coefficient) among operon and non-operon gene pairs. (E) Position specific mRNA folding energy mean margin between gene pairs in absolute values. The bars represent the position specific P-value (-log10pv) comparing the OPs mean value to that of NOPs. The scale for the bar charts is not given, instead the common significance threshold is drawn (see legend). All P-values were computed using an unsupervised standard permutation test (see ‘Materials and Methods’ section: Permutation test). All significant P-values were confirmed using the Benjamini & Hochberg False Discovery Rate (FDR) procedure (62), 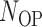 = 137,
= 137, 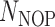 = 76.
= 76.