Figure 3.
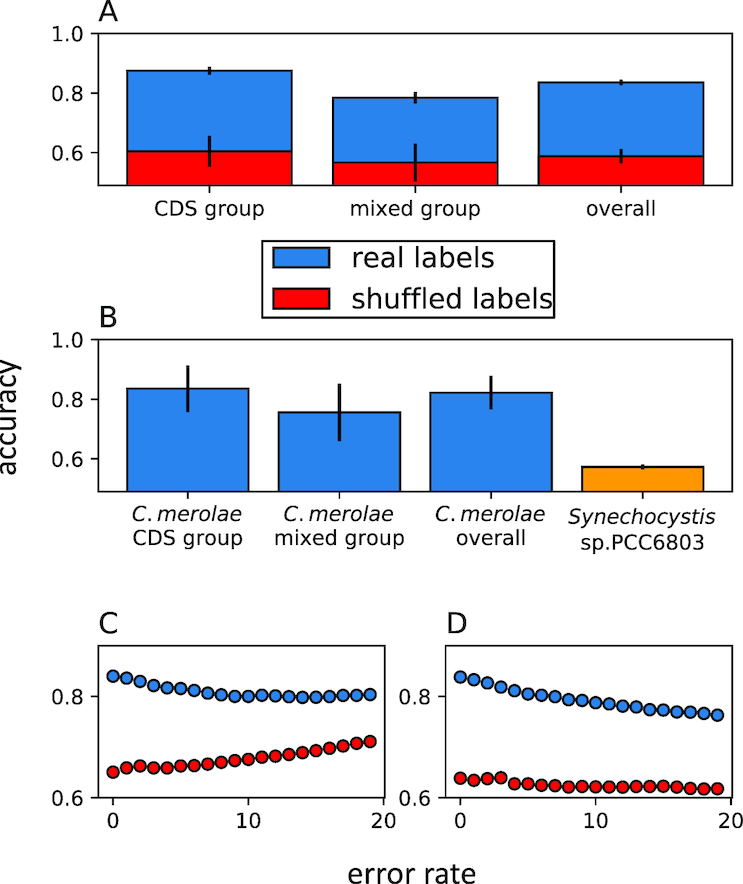
Model performance evaluation. (A) Accuracy scores of the random-forest classifier on real labels and random labels (see legend). 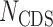 = 121,
= 121, 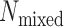 = 92. (B) Prediction accuracies on the red alga Cyanidioschyzon merolae when discarded from the initial dataset, and on the cyanobacteria Synechocystis sp. PCC6803 labels derived from (12). The bars represent coding-sequence gene pairs (‘CDS group’), a mixture of tRNA, rRNA or coding sequence gene pairs (‘mixed group’) and the weighted mean of the two groups (‘overall’). Synechocystis sp. PCC6803 encompasses only CDS gene pairs. The bars show mean ± STD. (C) Type I (false-positive mistake) error test. (D) type II (false-negative mistake) error test. An overall of 19 errors were introduced into the labels and for each error rate the prediction pipeline was repeated. The same analysis was carried out on random labels. All accuracy scores were calculated based on the average accuracies of ten bootstrap trained samples.
= 92. (B) Prediction accuracies on the red alga Cyanidioschyzon merolae when discarded from the initial dataset, and on the cyanobacteria Synechocystis sp. PCC6803 labels derived from (12). The bars represent coding-sequence gene pairs (‘CDS group’), a mixture of tRNA, rRNA or coding sequence gene pairs (‘mixed group’) and the weighted mean of the two groups (‘overall’). Synechocystis sp. PCC6803 encompasses only CDS gene pairs. The bars show mean ± STD. (C) Type I (false-positive mistake) error test. (D) type II (false-negative mistake) error test. An overall of 19 errors were introduced into the labels and for each error rate the prediction pipeline was repeated. The same analysis was carried out on random labels. All accuracy scores were calculated based on the average accuracies of ten bootstrap trained samples.
