Figure 5.
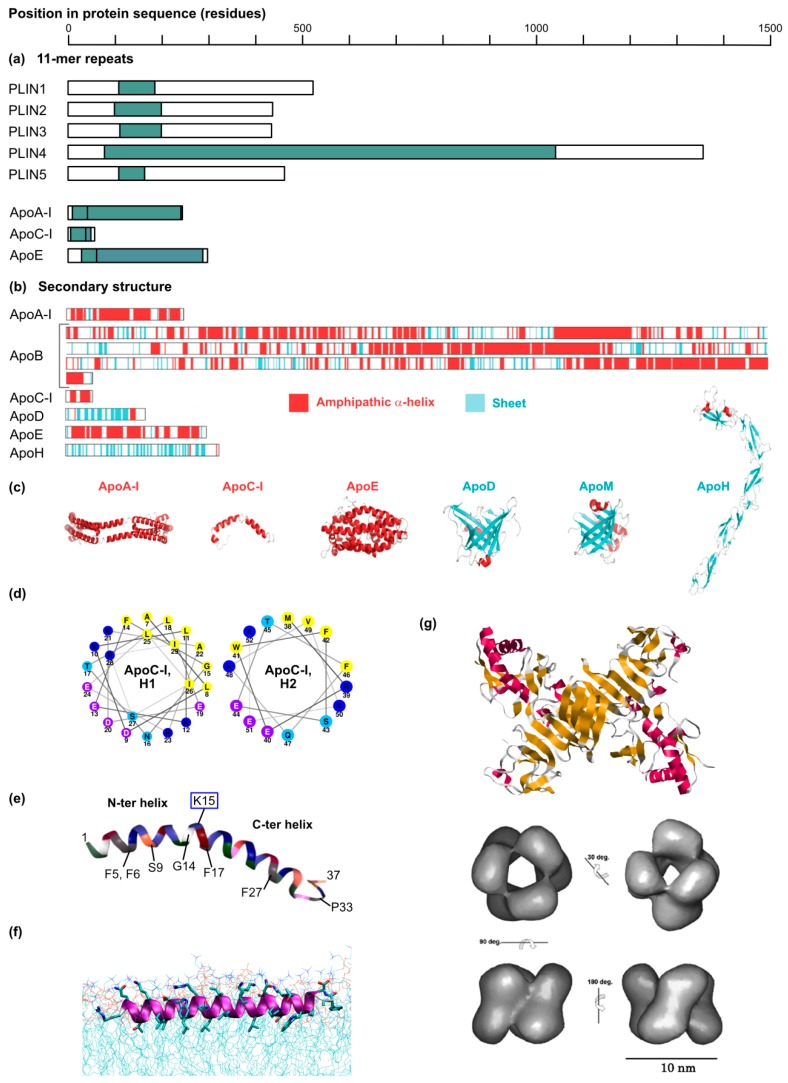
Striking sequence and structural features of apolipoproteins and apolipoprotein mimics. (a) Like the perilipins, apolipoproteins contain 11-mer repeats (teal-colored boxes) that have the propensity to fold as amphipathic α-helices. ApoA-I, C-I and E are shown as examples. For the apolipoproteins (the mature proteins are depicted), the two colored boxed correspond to 11-mer repeats found in exons 3 and 4, respectively. In some cases, the 11-mer repeats are organized themselves into larger motifs: 33-mer (3 × 11-mer) repeats for PLIN4 [154], 22-mer repeats for most 11-mers found in exon 4 of the apolipoproteins [155]. Note that PLIN2 is also called ADRP, and PLIN3 corresponds to TIP47. The coordinates of the 11-mer-containing domains have been extracted from Copic A. et al. [154] for the PLIN proteins and from Luo C.C. et al. [155] for the apolipoproteins. (b,c) Predicted or experimentally determined secondary structures and 3D models of a range of apolipoproteins. Note that ApoA-I, ApoB, ApoC-I and ApoE, which are predominantly folded as amphipathic α-helices, can rescue HCV virus production in 293T-derived cells but ApoD and H, which are mostly organized in sheets, cannot [82,117]. These two panels were reproduced and adapted from Fukuhara T. et al. [82] with permission of the authors. (d) Wheel representations of the two ApoC-I α-helices. The helices were drawn with Netwheels (http://lbqp.unb.br/NetWheels/) [58] and the color coding is as in Figure 2. (e) Structure of CAMP LL-37 domain in lipid micelles as determined by NMR (PDB ID: 2K6O, [139]) and visualized with RCSB PDB (www.rcsb.org) [156] and the NGL Viewer [157]. Key residues of the hydrophobic face apposed to the lipid micelles are indicated on the bottom. The lysine residue responsible for the hinge between the N- and C-terminal helices is indicated on the top and highlighted with a blue square. (f) Erns amphipathic α-helix (residues 194–227) reconstituted by molecular dynamics in a theoretical membrane. Reproduced from Aberle D. et al. [146] with permission of the authors. (g) Top panel: Crystal structure of the NS1 protein dimer of dengue virus type 2. Note the predominant sheet organization. Structure published in Akey D.L. et al. [149], PBD ID = 4O6B, visualized with Rastop, a software adapted from Rasmol (http://www.geneinfinity.org/rastop/) [158]. Bottom panel: 3D reconstruction of dengue virus 1 NS1 hexamer based on cryo-electron microscopy. The hexamer forms an open barrel of about 10 nm in diameter with a central channel (see top right image) believed to accommodate the neutral lipid core of this HDL-like particle. Reproduced from Gutsche et al. [151] with permission from the authors.