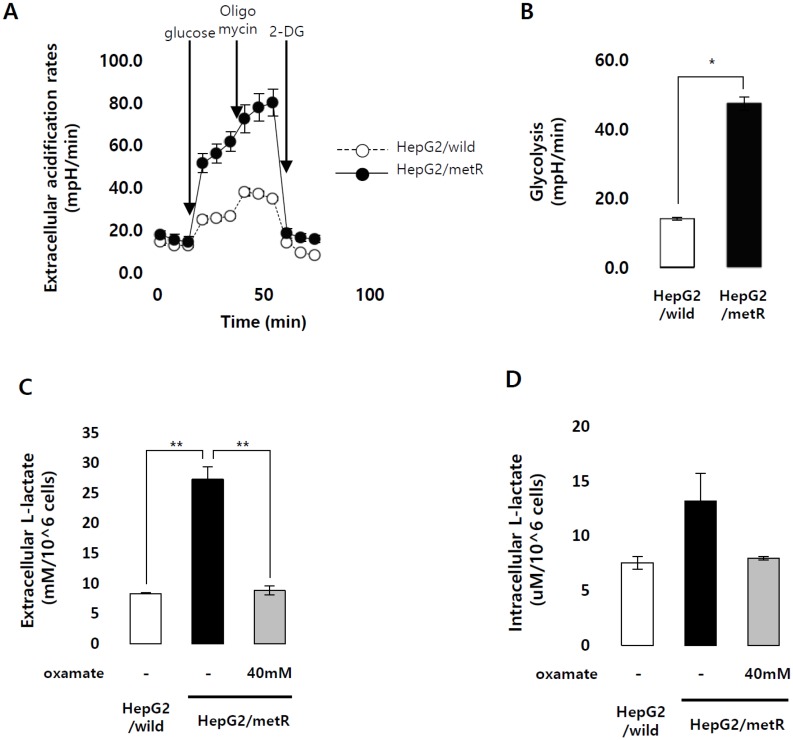
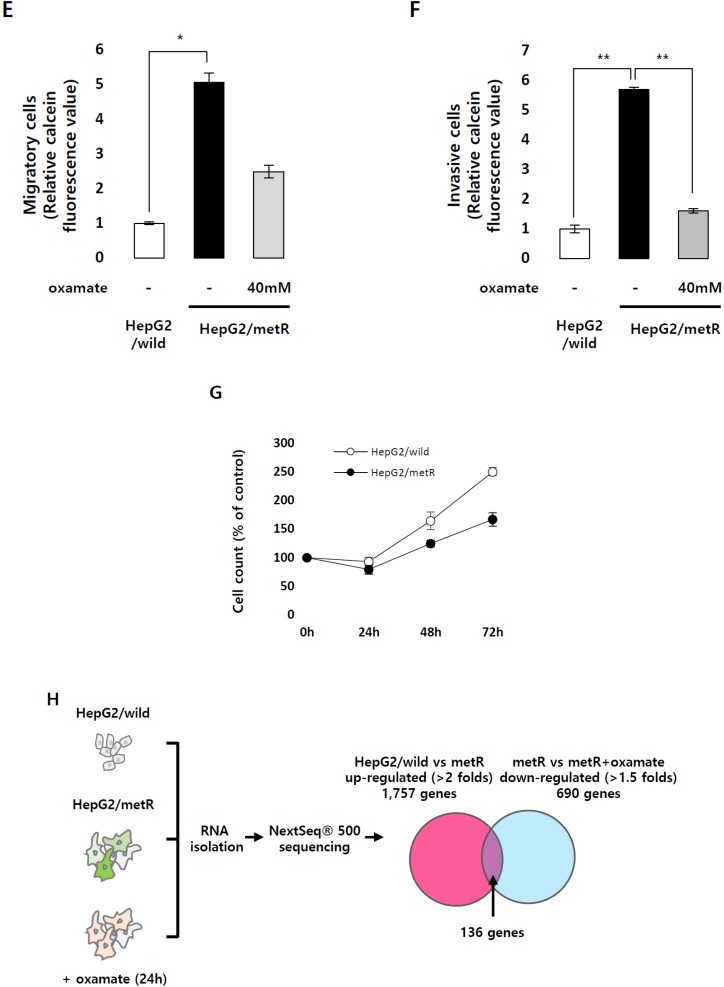
Figure 1.
Identification of genes differentially expressed in a lactate-enriched microenvironment. (A) Kinetic extracellular acidification rates (ECAR) response of parental HepG2 and HepG2/metR cells to glucose (20 mM), oligomycin (1 µM), and 2-DG (50 mM), respectively. ECAR values were normalized by the BCA protein assay. A representative experiment out of at least three trials is shown. (B) The glycolytic activity was measured by quantitatively estimating the conversion of glucose to pyruvate in HepG2 and HepG2/metR cells after the addition of a saturating amount of glucose to a given cell. p-value was assessed by Student’s t-test using data from triplicate experiments. * p < 0.05 and ** p < 0.01 vs. control. (C) Extracellular lactate production by HepG2, HepG2/metR, and oxamate-treated HepG2/metR cells. Results are mean ± SD of three experiments. The p-value was assessed by Student’s t-test. * p < 0.05 and ** p < 0.01 vs. control. (D) Intracellular lactate production by HepG2, HepG2/metR, and oxamate-treated HepG2/metR cells. Results are mean ± SD of three experiments. The p-value was assessed by Student’s t-test. (E,F). Migratory and invasive properties of HepG2, HepG2/metR, and oxamate-treated HepG2/metR cells as represented by relative calcein-AM fluorescence values. Results from three independent experiments are shown as mean ± SD. * p < 0.05 and ** p < 0.01 vs. control by Student’s t-test. (G) Growth of HepG2 and HepG2/metR cells were quantitatively estimated by cell counting during indicated time period. Results from three independent experiments are shown as means ± SD. (H) Schematic illustration of the RNA-sequencing strategy to profile lactate-induced gene expression.