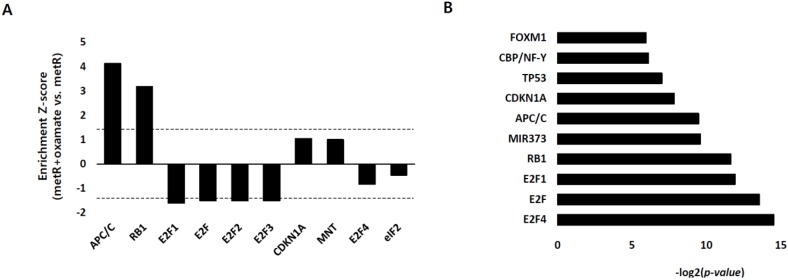
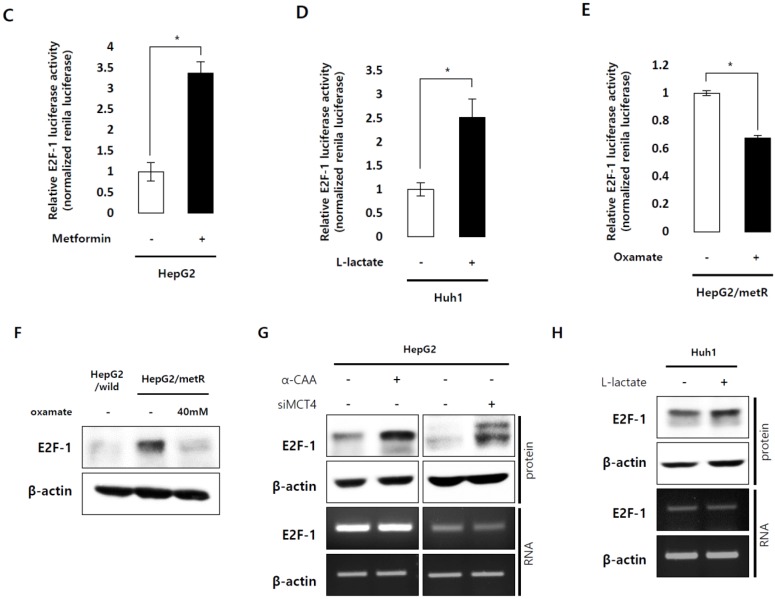
Figure 3.
Role of E2F1 pathway in lactate-induced gene expression and its regulation by lactate. (A) Profiling of upstream regulators of lactate-induced gene expression. Transcriptome data for HepG2/metR cells with or without oxamate treatment were subject to a sub-network enrichment analysis using Pathway Studio program, and the activation score (Z-score) of putative upstream regulators was quantitatively estimated. Dotted lines represent a statistical significance of p < 0.05 in both directions. (B) Upstream regulators predicted for the eleven lactate-responsive genes related to ‘microtubule functions’ through the literature mining process of Pathway Studio. (C–E). Lactate-dependent transcription activity of E2F1 was measured by E2F1-driven luciferase reporter assays in (C) metformin (10 mM)-treated HepG2 cells, (D) exogenous l-lactate (50 mM)-treated Huh1 cells, and (E) oxamate (40 mM)-treated HepG2/metR cells. The luciferase activity values were normalized by renilla luciferase values. All results from three independent experiments are shown as mean ± SD. The p-value was assessed by Student’s t-test. * p < 0.05 and ** p < 0.01 vs. control. (F–H). The protein and mRNA expression of E2F1 gene in various lactate-enriched microenvironments including (F) oxamate (40 mM)-treated HepG2/metR cells, (G) pharmacological inhibition of lactate transporter or siRNA-mediated depletion of MCT4 in HepG2 cells, and (H) exogenous l-lactate treatment in Huh1 cells.