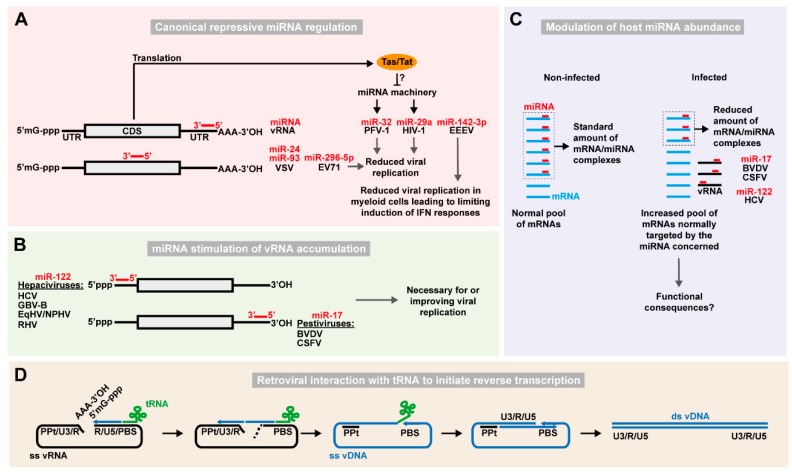
Figure 1.
Function of host ncRNAs directly interacting with viral RNA. (A) Examples of repressive regulation of RNA virus replication via canonical miRNA-dependent targeting. Host miRNAs (red bars) targeting viral RNA (vRNA) CDS or 3′ UTR are shown. Capped 5′-end (5′mG-ppp) and poly-adenylated 3′ end (AAA-3′OH) of the vRNA are also depicted. Viral proteins such as PFV-1 Tas or HIV-1 Tat may counteract the miRNA machinery. (B) Stimulation of RNA virus replication via miRNA-dependent targeting. Hepaciviruses and pestiviruses interact with miR-122 on the 5′ UTR and let-7 and miR-17 on the 3′ UTR, respectively, to stimulate IRES-driven translation, protection from degradation and replication leading to viral RNA accumulation. (C) Sponging of host miRNAs by viral RNA affects the expression of cellular mRNAs. In non-infected cells, a defined pool of miRNAs (red bars) targets the mRNA pool (light blue) for inhibition of translation or degradation. In infected cells, vRNA (black) can sponge a significant proportion of specific miRNA species resulting in a reduced available pool of that particular miRNA, and therefore a de-repression of normally targeted mRNAs. These effects have been described for the miRNA/vRNA combinations indicated. (D) Retroviral interaction with tRNAs to initiate reverse transcription of the viral RNA. In the Retroviridae family, a cellular tRNA (green) complementary to the primer binding site (PBS) sequence in the 5′ region of the viral (+)ssRNA (black) is used to initiate reverse transcription. The complementarity between the long terminal repeat sequences (including the unique 3′ [U3], repeat [R], and unique 5′ [U5] regions) at the ends of the vRNA assists in conversion of the viral genome into ssDNA (blue). Finally, synthesis of the second DNA strand is initiated from the polypurine tract (PPt) region that is resistant to RNAse H, degrading the remaining vRNA, to thereby generate the double-stranded vDNA (ds vDNA) that will get inserted into the cellular chromosome as a provirus.