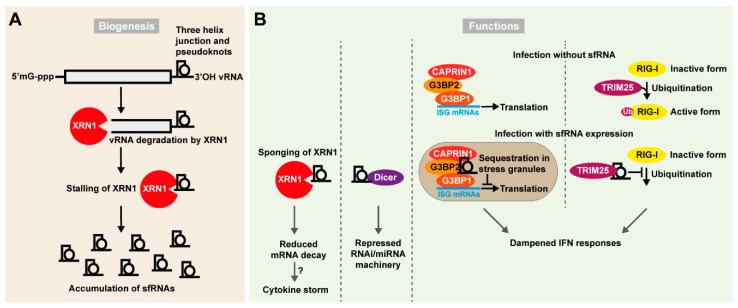
Figure 3.
Sub-genomic flavivirus RNAs (sfRNAs) impair cellular response to viral infection. (A) Biogenesis of sfRNAs. A pseudoknot-stabilized three junction helix is formed in the 3′ UTR of flaviviral genomes. This blocks the 5′ to 3′ processing and nucleolytic activity of the exonuclease XRN1, which triggers accumulation of sfRNAs in the cell. (B) Functions of sfRNAs. The accumulation of sfRNAs impacts several cellular pathways which can favor viral infection: in one mechanism, stalling at the sfRNA pseudoknots sequesters XRN1 away from its usual targets decreasing mRNA decay, including decay of transcripts involved in the antiviral response. This further could contribute to the cytokine storm observed upon infection. In another mechanism a high amount of sfRNAs saturates Dicer and consequently inhibits the RNAi/miRNA pathway. sfRNAs have also been reported to accumulate in stress granules and interact with CAPRIN1, G3BP2, and G3BP1 that are important for translation of ISG mRNAs. Finally, sfRNAs may interfere directly with the PRR mediated IFN-I activation pathway. RIG-I is a central component of PRR signaling and is activated by ubiquitination via TRIM25 upon infection. sfRNAs have been shown to directly bind TRIM25 and by doing so preventing the ubiquitination of RIG-I. This in turn would lead to a dampened IFN response.