Abstract
Since Ronald Konopka and Seymour Benzer’s discovery of the gene Period in the 1970s, the circadian rhythm field has diligently investigated regulatory mechanisms and intracellular transcriptional and translation feedback loops involving Period, and these investigations culminated in a 2017 Nobel Prize in Physiology or Medicine for Michael W. Young, Michael Rosbash, and Jeffrey C. Hall. Although research on 24-hour behavior rhythms started with Period, a series of discoveries in the past decade have shown us that post-transcriptional regulation and protein modification, such as phosphorylation and oxidation, are alternatives ways to building a ticking clock.
Keywords: circadian rhythms, post-transcriptional oscillator, transcriptional-translation feedback loop, red blood cells, dopaminergic ultradian oscillator, peroxiredoxin, phosphorylation
Introduction
The time-keeping mechanisms of circadian rhythms can be regulated by multiple layers of different cellular networks, including transcription-translation feedback loops (TTFLs) and post-translation oscillators (PTOs) 1. Circadian TTFLs generate oscillations in gene expression through delayed negative feedback whereby expression of a transcription factor negatively regulates its own transcription 2. The core of this genetic network in mammals is the expression of a heterodimer of BMAL1 (also called ARNTL) with either CLOCK or NPAS2, which binds at promoter cis-elements called E-boxes to drive expression of genes encoding period (PER1-3), cryptochrome (CRY1-2), and nuclear receptor subfamily (NR1D1-2) proteins, which then repress Bmal1 expression by a series of separate and interconnected feedback loops 3, 4. In contrast to behaviors driven by cyclic differences in gene expression, PTOs generate rhythms independent of transcription and translation through biochemical processes, such as phosphorylation, protein–protein interactions, and other post-translational modifications. These post-translational processes also alter TTFLs as well as post-transcriptional modification of transcripts involved in TTFLs 5, 6. The most well-known PTO is the cyanobacteria KaiABC system, which consists of only three proteins and ATP 7, but novel PTOs may also exist in red blood cells (RBCs) 8– 14, which lack a nucleus and the molecular machinery to drive TTFL rhythms. In addition, a series of new and old observations of 24-hour rhythms in biological contexts where classic TTFLs are absent or diminished ( Figure 1) 15– 23 continue to puzzle researchers and demonstrate that there are multiple ways to build a clock.
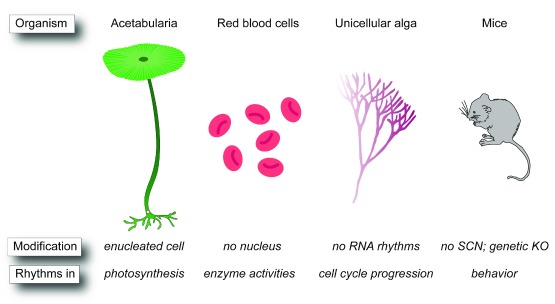
Figure 1. Post-translation oscillators without transcription-translation feedback loops.
Examples of post-translation oscillators in enucleated cells such as Acetabularia and red blood cells, in unicellular alga lacking RNA rhythms, and in mice in which the classic transcription-translation feedback loop module is disrupted genetically or anatomically. KO, knockout; SCN, suprachiasmatic nucleus.
One of the first modern uses of the term “circadian” was to describe 24-hour endogenous oscillators that alter Drosophila fly behavior rhythms 24, and the persistence of oscillations at various temperatures was viewed as a defining feature of circadian rhythms 25, 26. The first genetic component of circadian rhythms was discovered in the 1970s when Ronald Konopka in Seymour Benzer’s lab used chemical mutagenesis of Drosophila to discover three alleles of the Period gene 27. In the 1980s, rhythmicity of Period mutants was shown to be restored by gene transfer 28, 29, confirming that Period both is necessary and can restore rhythmic behaviors, such as eclosion and locomotor activity, in flies. In 1990, Hardin et al. proposed that PER protein altered the levels of Period mRNA in a negative feedback loop 30, but at the time it was unclear whether PER directly suppressed Period transcription or whether the negative feedback occurred through an indirect route. A few years later, researchers discovered that this negative feedback was direct in the bread mold Neurospora crassa model of circadian rhythms because the frequency (FRQ) directly repressed its own transcription 31. In addition to Neurospora 32 and Drosophila 33, TTFL models of circadian rhythms from plants 34 to mammals 35 have been elucidated and reviewed extensively.
Post-translational oscillators and post-translational modifications: breaking the transcription-translation feedback loop mold
The modern idea that TTFLs were necessary for 24-hour rhythms was shattered in 2005 when Nakajima et al. reconstituted rhythmic 24-hour oscillations in protein phosphorylation with just a small number of cyanobacterial proteins 36. This seminal moment in the circadian rhythm field spurred investigators to examine other non-canonical rhythm-generating mechanisms and to unearth forgotten studies of PTOs. For example, in the 1960s, it was shown that the unicellular alga Acetabularia undergoes diurnal rhythms of photosynthesis, which persist even after the nucleus has been artificially removed 21.
There are a few more recent examples of organisms that have circadian rhythms in the absence of TTFLs. In the unicellular red alga Cyanidioschyzon merolae, circadian rhythms control cell cycle progression in the absence of RNA translation 22, and the unicellular dinoflagellate Lingulodinium has daily rhythms in bioluminescence and photosynthesis without a detectable change in RNA transcript abundance and in the presence of transcription inhibitors 23. These studies suggest that protein activities and post-translational modifications can serve as 24-hour oscillators. Research has centered on phosphorylation as the period-determining post-translational modification 37– 40, but other post-translational modifications, including methylation, acetylation, sumoylation, and ubiquitination, also alter clock function 41– 44.
Importantly, circadian rhythms are insensitive to temperature and this property of temperature compensation was identified in biological time-keeping systems, such as those of bees, flies, and marine organisms, as early as the 1950s and 1960s 25, 26, 45, 46. Transcription and translation are temperature-dependent reactions 47– 50, which suggests that post-translational activities are important for temperature compensation. For example, Isojima et al. revealed that phosphorylation by casein kinase I (CKI) is a temperature-insensitive period-determining process, and the degradation rate of PER2, which is regulated by CKI phosphorylation, was found to be insensitive to temperature 38. Importantly, the phosphorylation of PER2-derived peptide by CKI is insensitive to temperature in vitro. In 2015, the degradation of PER2 was found to occur in a more complex mode composed of three distinct stages, and the duration of the second stage depended on circadian time, which led to the identification of temperature-sensitive and -insensitive PER2 phosphorylation sites 51. Thus, differences in the temperature sensitivity of phosphorylation sites on the repressor, which alter degradation rates at different temperatures, are responsible for temperature compensation, PER2 stability, and ultimately the length of the circadian period. In 2017, Shinohara et al. identified a short sequence region around residue K224 in CKI, which was responsible for temperature compensation and converted a temperature-sensitive kinase into a temperature-insensitive one in vitro 52. Mutation of K224 shortens circadian behavioral rhythms and alters the temperature dependency of the circadian clock in the sub-hypothalamic region of the brain 52, called the suprachiasmatic nucleus (SCN), which controls circadian response to light. It is though noteworthy that K224 is part of the consensus KRQK monopartite nuclear localization signal in CKI, which makes it difficult to disentangle the effects of temperature dependence from that of localization in vivo. These studies provide evidence for how post-translational activities modify TTFL rhythms, but a series of new and old studies have revealed that PTOs can drive rhythms even in the absence of TTFL clocks.
Blood: a novel source of post-translational oscillator rhythms
Mammals have a natural supply of enucleated cells in RBCs, and researchers have plumbed this cell type for non-TTFL rhythms. In the 1970s, circadian rhythms in ATPase activity and periodic rhythms in enzymes—such as acetylcholinesterase, glyceraldehyde-3-phosphate dehydrogenase, and glucose-6-phosphate dehydrogenase—in RBCs were found ( Table 1), but it was unclear whether the rhythms were robust or persistent beyond 24 hours 10.
Table 1. Oscillatory phenomena observed in human red blood cells.
| Molecule | Year | Period | Impact | Reference |
|---|---|---|---|---|
| Glucose-6-phosphate
dehydrogenase |
1975 | ~12 hours | Observed two peaks in enzyme activity over a 24-hour period in three
different individuals |
13 |
| Glutamate oxaloacetate
transaminase |
1975 | ~12 hours | Observed two peaks in enzyme activity over a 24-hour period in two
different individuals |
13 |
| Acid phosphatase | 1975 | ~24 hours | Observed one peak in enzyme activity over a 24-hour period in one
individual in plasma-free human red blood cell suspensions |
13 |
| Acetylcholinesterase | 1975 | ~24 hours | Observed one peak in enzyme activity over a 24-hour period in two
individuals |
13 |
| Glucose-6-phosphate
dehydrogenase |
1976 | ~12 hours | Observed two peaks in activity over a 24-hour period with one pattern
peaking at 4 p.m. and midnight and the other peaking at midnight and 8 p.m. in six and five individuals, respectively |
14 |
| 6-phophogluconate
dehydrogenase |
1976 | ~12 hours | Observed two peaks in activity over a 24-hour period peaking at
4 a.m. and 4 p.m. in 11 individuals |
14 |
| Lactic dehydrogenase | 1976 | ~12 hours | Observed two peaks in activity over a 24-hour period with one pattern
peaking at noon and midnight and the other peaking at 4 a.m. and 4 p.m. in four and seven individuals, respectively |
14 |
| Aspirate aminotransferase | 1976 | ~12 hours | Observed two peaks in activity over a 24-hour period peaking at 4 a.m.
and 4 p.m. in 11 individuals |
14 |
| Hexokinase | 1976 | ~24 hours | Observed one peak in activity over a 24-hour period peaking at 4 p.m.
in 11 individuals |
14 |
| Potassium efflux | 1976 | NS | Observed a steady increase in potassium efflux over a 48-hour period in
an unknown number of individuals (averaged data reported) |
12 |
| Membrane potential | 1976 | ~24 hours | Observed two peaks in membrane potential by DiOC
5(3) over a 48-hour
period in an unknown number of individuals (averaged data reported) |
12 |
| Mg-dependent ATPase | 1976 | ~24 hours | Observed one peak in activity from human blood bank bags incubated
at 37 °C for 27 hours (average of eight samples) |
8 |
| Acetylcholinesterase | 1978 | NS | Observed variations in acetylcholinesterase activity over a 24-hour
period in four individuals, but variations had lower amplitude than reference 13 and were not circadian |
10 |
| Peroxiredoxin | 2011 | ~24 hours | Observed three peaks of peroxiredoxin dimer oxidation and PRX-SO
2/3
abundance over a 60-hour period in three individuals |
11 |
| NADH | 2011 | ~24 hours | Observed three peaks in NADH abundance over a 60-hour period in
three individuals |
11 |
| NADPH | 2011 | ~24 hours | Observed three peaks in NADPH abundance over a 60-hour period in
three individuals |
11 |
| Membrane potential | 2017 | ~24 hours | Observed two peaks in membrane potential by dielectrophoresis,
DiOC 5(3), and mass spectrometry over a 48-hour period in four biological replicates |
9 |
| Membrane conductance and
cytoplasm conductivity |
2017 | ~24 hours | Observed two or three peaks in membrane conductance and cytoplasm
conductivity over a 48-hour period in four individuals |
9 |
| Intracellular potassium | 2017 | ~24 hours | Observed two peaks in intracellular potassium concentrations over a
48-hour period in four biological replicates |
9 |
NS, not significant.
In 2011, an anti-oxidant enzyme called peroxiredoxin (PRX) in cultured human RBCs was found to have temperature-independent circadian cycles of hyperoxidation for up to 76 hours 11. Because RBCs lack a nucleus and the rhythms persisted in the presence of transcription and translation inhibitors, a novel non-transcriptional-based circadian oscillator in mammals was proposed. Analysis of the PRX rhythms relied solely on PRX1, PRX2, and PRX-SO 2/3 (hyperoxidized PRX form) antibodies. In particular, the PRX-SO 2/3 antibody recognizes multiple hyperoxidized forms of PRX 53, results in up to eight different bands on non-reducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) 11, and produces multiple non-specific bands that can confound interpretation of the hyperoxidized signal 54, 55, which make determination of the correct PRX isoform technically difficult and in-gel controls essential. Nevertheless, the same researchers discovered that hyperoxidized PRX-SO 2/3 rhythms were conserved in a wide range of species 55, 56.
In mice, blocking hemoglobin oxygen transport by incubation with carbon monoxide eliminates PRX2 hyperoxidized rhythms 57. Hemoglobin auto-oxidation in RBCs generates superoxide, which is converted to H 2O 2 by superoxide dismutase 1 (SOD1) 58, 59, and H 2O 2 is subsequently reduced by catalase, glutathione peroxidase, and PRXs 54, 58, 60, which results in the oxidation of these proteins 61. Oxidation of PRX2 is reversed by sulfiredoxin (SRX) 62– 64, but rhythms in PRX2 oxidation in mice are not mediated by the rhythmic reduction of hyperoxidized PRX2 by SRX but rather through rhythmic degradation by 20S proteasomes, and only about 1% of the total PRX pool is modified in a circadian manner 57. Mitochondria-specific PRX (PRX3) is also reversibly inactivated by hyperoxidation, reduced, and reactivated by SRX, and hyperoxidized PRX3 and SRX undergo anti-phasic circadian oscillations in the mitochondria in various mice tissues, which links mitochondria function to circadian rhythms 65. Another group revealed about three peaks in hyperoxidized PRX-SO 2/3 rhythms in mice over a 48-hour period (instead of two as would be expected for a circadian rhythm) and showed that rhythms were impaired in SOD1-mutant mice 66. There is still uncertainty regarding the origin of PRX hyperoxidation rhythms, but the data suggest that PRX-SO 2/3 oscillations are more an output of rhythm-generating machinery involving the 20S proteasome rather than a daily oxidation-reduction cycle.
Deconstructing PRX rhythms biochemically and using non-antibody methods, such as mass spectrometry, to directly detect the hyperoxidized cysteine residue or redox-sensitive fluorescent proteins may bolster understanding of this novel PTO. However, biochemical reconstruction is difficult because RBC lysis causes gradual loss of PRX-SO 2/3 signal over a 48-hour period 9. However, these types of approaches have revealed that potassium-containing media enhances PRX2-SO 2/3 rhythms 9, and chemical perturbation with Conoidin A, a PRX2 inhibitor 67, shortens PER2:LUCIFERASE rhythms in immortalized mouse fibroblasts 68.
The exact mechanism of rhythmic PRX oxidation is still unclear, but researchers have begun to examine other general rhythmic behaviors in RBCs. Although no one has followed up on the circadian, ultradian, and irregular rhythms of various enzyme activities in the 1970s, in 2011 researchers reported circadian changes in NADH and NADPH levels 11, and in 2017 researchers reproduced circadian changes in RBC membrane potential 9 observed in an article published in 1976 12. Paradoxically, the researchers reported circadian changes in potassium concentration in 2017, whereas no rhythms in potassium were observed in 1976; instead, a gradual and steady increase in potassium efflux occurred over the 48-hour observation period. Whether these differences arise from measuring slightly different potassium populations (intracellular versus extracellular), a small sample size, technical differences in methods and individuals, or an actual biological phenomenon remains to be determined.
Post-translation control of circadian period in transcription-translation feedback loop model organisms
There are shared design principles between the period-determination processes of PTO-based and TTFL-based oscillators. A PTO generates rhythmic changes in protein states without changing the amount of protein itself. On the other hand, rhythmic protein synthesis and degradation are essential for TTFL-based oscillations, and mechanisms that control protein abundance are critical for controlling the circadian period. This idea is widely accepted for circadian TTFL oscillators because there is a significant correlation between the half-life of transcription repressor mutants, such as in Drosophila PER and Neurospora FRQ, and circadian period 69, 70. The correlation suggests that faster degradation of circadian repressors accelerates clock speed. In mammalian circadian clocks, F-box proteins recruit E3 ubiquitin ligase complexes that license PER and CRY degradation, which modulates period length 71. Although the circadian TTFL-based oscillators involve post-translational regulation as period-determination mechanisms, modification of transcription repressors regulates period length by changing repressor stability. For example, a mutation in CKIε that destabilizes mammalian PER results in period shortening 72, 73, and mutation of a phosphorylation site on PER that destabilizes PER also results in period shortening 74, 75. Other kinases, such as AMPK and DNA-PK, control period length by altering CRY stability through phosphorylation 76, 77. In addition, stabilization of CRY by small molecules lengthens the period 78, and destabilization of CRY by degron tagging of CRY shortens the period 79, strongly suggesting the causal relationship between CRY stability and period length.
However, a recent study of the Neurospora circadian clock challenged this protein stability–period length paradigm of period determination in a TTFL-based oscillator 80. Researchers used an FWD-1–deleted strain, which is an F-box protein that causes proteolysis of phosphorylated FRQ. The Δfrd-1 strain results in a markedly increased FRQ half-life, and new FRQ is produced even in the presence of hyperphosphorylated FRQ. Nonetheless, circadian oscillation of FRQ-promoter activities persists with modest change in period length, and several short-period mutations of FRQ still have a short period in a Δfrd-1 background in which the stability of FRQ is significantly increased. Because mutation of phosphorylation sites in FRQ still alters the period and because generic inhibition of kinase activity lengthens the period even in the absence of FWD-1, these data suggest that a protein-state not a protein-abundance attribute, namely phosphorylation, controls period length.
A similar uncoupling of protein stability and circadian period may occur even in the TTFL clock in mammals. A recent study of CRY1 mutations in phosphorylation sites by Ode et al. revealed that multiple phosphorylation sites near the co-factor binding pocket of CRY1 markedly changes period length while having only a modest effect on CRY1 half-life 79. Mutagenesis of CRY1 and CRY2 revealed mutations in a secondary co-factor binding pocket which shorten the period without reducing CRY1 stability 81. Furthermore, an exon-skipping mutation in CRY1 found from a human family with delayed sleep phase syndrome lengthens the period without affecting CRY1 stability 82. Therefore, mammalian CRY may also control the circadian period independently of its abundance.
If protein abundance control does not explain all aspects of period determination, what is the nature of state control of TTFL-based oscillator proteins such as multisite phosphorylation of FRQ, PER, and CRY? One of the shared properties of period-determining repressor proteins is structural flexibility. Most FRQ and PER regions modified by multisite phosphorylation are intrinsically disordered, highly flexible, and variable 83, 84. The multisite phosphorylation region of CRY1 critical for period control also occurs on a flexible loop region. These flexible regions may undergo a relatively large conformation change that may underlie slow dynamics (that is, 24 hours) of protein activity change. The intrinsically disordered C-terminal domain of BMAL1 controls the period through a slow conformation change with a high energy barrier 85. Conformation changes may lead to a slow and coherent re-organization of the macromolecular repressor complex 86, which is consistent with the dynamics of the cyanobacteria PTO 87, 88 in which the slow dynamics of the intrinsic conformational change of KaiC 89 couple to the re-organization of the KaiABC complex 90. An atomic-scale understanding of the repressor complex in a TTFL-based oscillator may reveal subtle differences in molecular mechanisms of 24-hour period determination between PTO- and TTFL-based oscillators.
Oscillations without classic transcription-translation feedback loop oscillators
Several classic models of circadian rhythms have persistent 24-hour rhythms even when the circadian TTFL machinery is absent or disrupted. In S2 cells, which are generally regarded as non-rhythmic, a multi-omics approach recently revealed hundreds of genes, proteins, and metabolites with 24-hour rhythms 20. Although this approach seems to suggest the presence of a novel non-canonical oscillator with 24-hour periodicity, it does not preclude possible cell cycle effects from the roughly 24-hour doubling time of S2 cells or the possibility of classic circadian clock components operating below the experimental limits of detection. For example, large-scale proteomics studies of circadian variation frequently fail to detect circadian proteins 91, 92 because there may be only a few hundred to a thousand protein copies per cell 93. Thus, genetic knockout (KO) of canonical clock genes is needed to definitively determine whether rhythms derive from a novel oscillator.
In mammals, genetic and anatomical ablation of the circadian machinery normally disrupts 24-hour behavioral rhythms, but rhythms persist under specialized situations. For example, SCN-lesioned rats administered methamphetamine in the drinking water retain circadian behaviors of activity in constant light conditions 17. This so-called methamphetamine-sensitive oscillator also does not depend on classic circadian genes, such as Per1-2, Cry1-2, Bmal1, Npas2, and Clock 18, 94. Recent data suggest that the methamphetamine-sensitive oscillator is a long-period manifestation of a tunable dopamine ultradian oscillator 95, 96. KO of a dopamine transporter in SCN-lesioned or Bmal1 KO mice, which prevents dopamine reuptake in dopaminergic neurons, increases the period of the ultradian rhythms. Similarly, administration of methamphetamine, which increases extracellular dopamine concentrations, lengthens ultradian rhythms in a dose-dependent manner from 4 hours to an astonishing 48 hours. In contrast, the anti-psychotic drug haloperidol, which selectively blocks the dopamine D2 receptor, shortens long-period rhythms induced by methamphetamine in wild-type and Bmal1 KO mice 95. These data suggest that dopamine neurons are a second independent rhythm-generating mechanism in the brain, and future studies using chemical and genetic approaches to perturb dopamine pathways coupled with recently developed brain-clearing techniques 97– 100 may enable a more complete understanding of the neural architecture of this dopamine ultradian oscillator.
Conclusions
From blood to brain, these studies suggest that non-canonical PTOs have an impact on circadian rhythms beyond the classic PER negative feedback loop. However, recent studies of PER itself, including temperature-sensitive phosphorylation sites 51, three prime untranslated region (3′-UTR) regulation 101, and the separation of Period2 rhythms from Bmal1 rhythms in the SCN 102, indicate that even a gene as well studied as Period can still teach us new tricks about the period-determining mechanisms of circadian rhythms.
Abbreviations
AMPK, AMP-activated protein kinase; BMAL1, brain and muscle Arnt-like protein 1; CKI, casein kinase I; CLOCK, circadian locomotor output cycles kaput; CRY1-2, cryptochrome1-2; FRQ, frequency; FWD-1, F-box/WD-40-repeat-containing protein 1; KO, knockout; NPAS2, neuronal PAS domain-containing protein 2; NR1D1-2; nuclear receptor subfamily 1 group D member 1-2; PER; period; PRX, peroxiredoxin; PTO, post-translational oscillator; RBC, red blood cell; S2, Schneider 2; SCN, suprachiasmatic nucleus; SOD1, superoxide dismutase 1; SRX, sulfiredoxin; TTFL, transcription-translation feedback loop.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Carrie Partch, Department of Chemistry and Biochemistry, University of California Santa Cruz, Santa Cruz, CA, USA
John O'Neill, MRC Laboratory of Molecular Biology, Cambridge, UK
Funding Statement
This work was supported by a grant from the Japan Agency for Medical Research and Development (AMED)-Core Research for Evolution Science and Technology (CREST) (JP17gm0610006; AMED/Ministry of Education, Culture, Sports, Science and Technology [MEXT]; to HRU), CREST (Japan Science and Technology Agency/MEXT; to HRU), Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS) (AMED/MEXT; to HRU), Basic Science and Platform Technology Program for Innovative Biological Medicine (AMED/MEXT; to HRU), a Grant-in-Aid for Scientific Research (Japan Society for the Promotion of Science 18H05270 to HRU), a Grant-in-Aid for Scientific Research on Innovative Areas (23115006 to HRU), the strategic programs for research and development (President’s Discretionary Fund) of RIKEN (to HRU), an intramural Grant-in-Aid from the RIKEN Quantitative Biology Center (to HRU), a Grant-in-Aid for Young Scientists (18K14683 to KLO), the RIKEN Special Postdoctoral Research Program (to AM), and a Grant-in-Aid for Early-Career Scientists (18K14755 to AM).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved]
References
- 1. Hurley JM, Loros JJ, Dunlap JC: Circadian Oscillators: Around the Transcription-Translation Feedback Loop and on to Output. Trends Biochem Sci. 2016;41(10):834–46. 10.1016/j.tibs.2016.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Partch CL, Green CB, Takahashi JS: Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24(2):90–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown SA, Kowalska E, Dallmann R: (Re)inventing the circadian feedback loop. Dev Cell. 2012;22(3):477–87. 10.1016/j.devcel.2012.02.007 [DOI] [PubMed] [Google Scholar]
- 4. Ueda HR, Hayashi S, Chen W, et al. : System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37(2):187–92. 10.1038/ng1504 [DOI] [PubMed] [Google Scholar]
- 5. Kojima S, Green CB: Circadian genomics reveal a role for post-transcriptional regulation in mammals. Biochemistry. 2015;54(2):124–33. 10.1021/bi500707c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kojima S, Shingle DL, Green CB: Post-transcriptional control of circadian rhythms. J Cell Sci. 2011;124(Pt 3):311–20. 10.1242/jcs.065771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Swan JA, Golden SS, LiWang A, et al. : Structure, function, and mechanism of the core circadian clock in cyanobacteria. J Biol Chem. 2018;293(14):5026–34. 10.1074/jbc.TM117.001433 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 8. Cornelius G, Rensing L: Daily rhythmic changes in Mg 2+-dependent ATPase activity in human red blood cell membranes in vitro. Biochem Biophys Res Commun. 1976;71(4):1269–72. 10.1016/0006-291X(76)90791-9 [DOI] [PubMed] [Google Scholar]
- 9. Henslee EA, Crosby P, Kitcatt SJ, et al. : Rhythmic potassium transport regulates the circadian clock in human red blood cells. Nat Commun. 2017;8(1): 1978. 10.1038/s41467-017-02161-4 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 10. Mabood SF, Newman PF, Nimmo IA: Circadian rhythms in the activity of acetylcholinesterase of human erythrocytes incubated in vitro [proceedings]. Biochem Soc Trans. 1978;6(1):305–8. 10.1042/bst0060305 [DOI] [PubMed] [Google Scholar]
- 11. O'Neill JS, Reddy AB: Circadian clocks in human red blood cells. Nature. 2011;469(7331):498–503. 10.1038/nature09702 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 12. Hartman H, Ashkenazi I: Circadian changes in membrane properties of human red blood cells in vitro, as measured by a membrane probe. FEBS Lett. 1976;67(2):161–3. 10.1016/0014-5793(76)80356-0 [DOI] [PubMed] [Google Scholar]
- 13. Ashkenazi IE, Hartman H, Strulovitz B, et al. : Activity rhythms of enzymes in human red blood cell suspensions. J Interdiscipl Cycle Res. 1975;6(4):291–301. 10.1080/09291017509359494 [DOI] [Google Scholar]
- 14. Brok-Simoni F, Ashkenazi YE, Ramot B, et al. : The diurnal rhythm of enzymes in human red cells. Br J Haematol. 1976;32(4):601–8. 10.1111/j.1365-2141.1976.tb00964.x [DOI] [PubMed] [Google Scholar]
- 15. Clarke JD, Coleman GJ: Persistent meal-associated rhythms in SCN-lesioned rats. Physiol Behav. 1986;36(1):105–13. 10.1016/0031-9384(86)90082-X [DOI] [PubMed] [Google Scholar]
- 16. Dragovic Z, Tan Y, Görl M, et al. : Light reception and circadian behavior in 'blind' and 'clock-less' mutants of Neurospora crassa. EMBO J. 2002;21(14):3643–51. 10.1093/emboj/cdf377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Honma K, Honma S, Hiroshige T: Activity rhythms in the circadian domain appear in suprachiasmatic nuclei lesioned rats given methamphetamine. Physiol Behav. 1987;40(6):767–74. 10.1016/0031-9384(87)90281-2 [DOI] [PubMed] [Google Scholar]
- 18. Mohawk JA, Baer ML, Menaker M: The methamphetamine-sensitive circadian oscillator does not employ canonical clock genes. Proc Natl Acad Sci U S A. 2009;106(9):3519–24. 10.1073/pnas.0813366106 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 19. de Paula RM, Lewis ZA, Greene AV, et al. : Two circadian timing circuits in Neurospora crassa cells share components and regulate distinct rhythmic processes. J Biol Rhythms. 2006;21(3):159–68. 10.1177/0748730406288338 [DOI] [PubMed] [Google Scholar]
- 20. Rey G, Milev NB, Valekunja UK, et al. : Metabolic oscillations on the circadian time scale in Drosophila cells lacking clock genes. Mol Syst Biol. 2018;14(8):e8376. 10.15252/msb.20188376 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 21. Sweeney BM, Haxo FT: Persistence of a Photosynthetic Rhythm in Enucleated Acetabularia. Science. 1961;134(3487):1361–3. 10.1126/science.134.3487.1361 [DOI] [PubMed] [Google Scholar]
- 22. Miyagishima Sy, Fujiwara T, Sumiya N, et al. : Translation-independent circadian control of the cell cycle in a unicellular photosynthetic eukaryote. Nat Commun. 2014;5: 3807. 10.1038/ncomms4807 [DOI] [PubMed] [Google Scholar]
- 23. Roy S, Beauchemin M, Dagenais-Bellefeuille S, et al. : The Lingulodinium circadian system lacks rhythmic changes in transcript abundance. BMC Biol. 2014;12:107. 10.1186/s12915-014-0107-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pittendrigh CS: Circadian rhythms and the circadian organization of living systems. Cold Spring Harb Symp Quant Biol. 1960;25:159–84. 10.1101/SQB.1960.025.01.015 [DOI] [PubMed] [Google Scholar]
- 25. Hastings JW, Sweeney BM: On the Mechanism of Temperature Independence in a Biological Clock. Proc Natl Acad Sci U S A. 1957;43(9):804–11. 10.1073/pnas.43.9.804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pittendrigh CS: On Temperature Independence in the Clock System Controlling Emergence Time in Drosophila. Proc Natl Acad Sci U S A. 1954;40(10):1018–29. 10.1073/pnas.40.10.1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Konopka RJ, Benzer S: Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68(9):2112–6. 10.1073/pnas.68.9.2112 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 28. Bargiello TA, Jackson FR, Young MW: Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature. 1984;312(5996):752–4. 10.1038/312752a0 [DOI] [PubMed] [Google Scholar]
- 29. Zehring WA, Wheeler DA, Reddy P, et al. : P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell. 1984;39(2 Pt 1):369–76. 10.1016/0092-8674(84)90015-1 [DOI] [PubMed] [Google Scholar]
- 30. Hardin PE, Hall JC, Rosbash M: Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343(6258):536–40. 10.1038/343536a0 [DOI] [PubMed] [Google Scholar]
- 31. Aronson BD, Johnson KA, Loros JJ, et al. : Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science. 1994;263(5153):1578–84. 10.1126/science.8128244 [DOI] [PubMed] [Google Scholar]
- 32. Dunlap JC, Loros JJ: Making Time: Conservation of Biological Clocks from Fungi to Animals. Microbiol Spectr. 2017;5(3). 10.1128/microbiolspec.FUNK-0039-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 33. Dubowy C, Sehgal A: Circadian Rhythms and Sleep in Drosophila melanogaster. Genetics. 2017;205(4):1373–97. 10.1534/genetics.115.185157 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 34. Nohales MA, Kay SA: Molecular mechanisms at the core of the plant circadian oscillator. Nat Struct Mol Biol. 2016;23(12):1061–9. 10.1038/nsmb.3327 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 35. Takahashi JS: Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18(3):164–79. 10.1038/nrg.2016.150 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 36. Nakajima M, Imai K, Ito H, et al. : Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308(5720):414–5. 10.1126/science.1108451 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 37. Más P, Kim WY, Somers DE, et al. : Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003;426(6966):567–70. 10.1038/nature02163 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 38. Isojima Y, Nakajima M, Ukai H, et al. : CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci U S A. 2009;106(37):15744–9. 10.1073/pnas.0908733106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chiu JC, Ko HW, Edery I: NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell. 2011;145(3):357–70. 10.1016/j.cell.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee HM, Chen R, Kim H, et al. : The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc Natl Acad Sci U S A. 2011;108(39):16451–6. 10.1073/pnas.1107178108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brown SA, Ripperger J, Kadener S, et al. : PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308(5722):693–6. 10.1126/science.1107373 [DOI] [PubMed] [Google Scholar]
- 42. Etchegaray JP, Lee C, Wade PA, et al. : Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421(6919):177–82. 10.1038/nature01314 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 43. Cardone L, Hirayama J, Giordano F, et al. : Circadian clock control by SUMOylation of BMAL1. Science. 2005;309(5739):1390–4. 10.1126/science.1110689 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 44. Naidoo N, Song W, Hunter-Ensor M, et al. : A role for the proteasome in the light response of the timeless clock protein. Science. 1999;285(5434):1737–41. 10.1126/science.285.5434.1737 [DOI] [PubMed] [Google Scholar]
- 45. Bruce VG, Pittendrigh CS: Temperature Independence in a Unicellular "Clock". Proc Natl Acad Sci U S A. 1956;42(9):676–82. 10.1073/pnas.42.9.676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Enright JT: Temperature compensation in short-duration time-measurement by an intertidal amphipod. Science. 1967;156(3781):1510–2. 10.1126/science.156.3781.1510 [DOI] [PubMed] [Google Scholar]
- 47. Oliveira SM, Häkkinen A, Lloyd-Price J, et al. : Temperature-Dependent Model of Multi-step Transcription Initiation in Escherichia coli Based on Live Single-Cell Measurements. PLoS Comput Biol. 2016;12(10):e1005174. 10.1371/journal.pcbi.1005174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Abbondanzieri EA, Shaevitz JW, Block SM: Picocalorimetry of transcription by RNA polymerase. Biophys J. 2005;89(6):L61–3. 10.1529/biophysj.105.074195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Farewell A, Neidhardt FC: Effect of temperature on in vivo protein synthetic capacity in Escherichia coli. J Bacteriol. 1998;180(17):4704–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Craig N: Effect of reduced temperatures on protein synthesis in mouse L cells. Cell. 1975;4(4):329–35. 10.1016/0092-8674(75)90153-1 [DOI] [PubMed] [Google Scholar]
- 51. Zhou M, Kim JK, Eng GW, et al. : A Period2 Phosphoswitch Regulates and Temperature Compensates Circadian Period. Mol Cell. 2015;60(1):77–88. 10.1016/j.molcel.2015.08.022 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 52. Shinohara Y, Koyama YM, Ukai-Tadenuma M, et al. : Temperature-Sensitive Substrate and Product Binding Underlie Temperature-Compensated Phosphorylation in the Clock. Mol Cell. 2017;67(5):783–798.e20. 10.1016/j.molcel.2017.08.009 [DOI] [PubMed] [Google Scholar]
- 53. Cox AG, Pearson AG, Pullar JM, et al. : Mitochondrial peroxiredoxin 3 is more resilient to hyperoxidation than cytoplasmic peroxiredoxins. Biochem J. 2009;421(1):51–8. 10.1042/BJ20090242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Low FM, Hampton MB, Peskin AV, et al. : Peroxiredoxin 2 functions as a noncatalytic scavenger of low-level hydrogen peroxide in the erythrocyte. Blood. 2007;109(6):2611–7. 10.1182/blood-2006-09-048728 [DOI] [PubMed] [Google Scholar]
- 55. O'Neill JS, van Ooijen G, Dixon LE, et al. : Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469(7331):554–8. 10.1038/nature09654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Edgar RS, Green EW, Zhao Y, et al. : Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485(7399):459–64. 10.1038/nature11088 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 57. Cho CS, Yoon HJ, Kim JY, et al. : Circadian rhythm of hyperoxidized peroxiredoxin II is determined by hemoglobin autoxidation and the 20S proteasome in red blood cells. Proc Natl Acad Sci U S A. 2014;111(33):12043–8. 10.1073/pnas.1401100111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Johnson RM, Goyette G, Jr, Ravindranath Y, et al. : Hemoglobin autoxidation and regulation of endogenous H 2O 2 levels in erythrocytes. Free Radic Biol Med. 2005;39(11):1407–17. 10.1016/j.freeradbiomed.2005.07.002 [DOI] [PubMed] [Google Scholar]
- 59. Winterbourn CC: Free-radical production and oxidative reactions of hemoglobin. Environ Health Perspect. 1985;64:321–30. 10.1289/ehp.8564321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cho CS, Lee S, Lee GT, et al. : Irreversible inactivation of glutathione peroxidase 1 and reversible inactivation of peroxiredoxin II by H 2O 2 in red blood cells. Antioxid Redox Signal. 2010;12(11):1235–46. 10.1089/ars.2009.2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rhee SG, Chae HZ, Kim K: Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38(12):1543–52. 10.1016/j.freeradbiomed.2005.02.026 [DOI] [PubMed] [Google Scholar]
- 62. Jeong W, Park SJ, Chang TS, et al. : Molecular mechanism of the reduction of cysteine sulfinic acid of peroxiredoxin to cysteine by mammalian sulfiredoxin. J Biol Chem. 2006;281(20):14400–7. 10.1074/jbc.M511082200 [DOI] [PubMed] [Google Scholar]
- 63. Biteau B, Labarre J, Toledano MB: ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425(6961):980–4. 10.1038/nature02075 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Woo HA, Chae HZ, Hwang SC, et al. : Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science. 2003;300(5619):653–6. 10.1126/science.1080273 [DOI] [PubMed] [Google Scholar]
- 65. Rhee SG, Kil IS: Mitochondrial H 2O 2 signaling is controlled by the concerted action of peroxiredoxin III and sulfiredoxin: Linking mitochondrial function to circadian rhythm. Free Radic Biol Med. 2016;100:73–80. 10.1016/j.freeradbiomed.2016.10.011 [DOI] [PubMed] [Google Scholar]
- 66. Homma T, Okano S, Lee J, et al. : SOD1 deficiency induces the systemic hyperoxidation of peroxiredoxin in the mouse. Biochem Biophys Res Commun. 2015;463(4):1040–6. 10.1016/j.bbrc.2015.06.055 [DOI] [PubMed] [Google Scholar]
- 67. Haraldsen JD, Liu G, Botting CH, et al. : Identification of Conoidin a as a Covalent Inhibitor of Peroxiredoxin II. Org Biomol Chem. 2009;7:3040–8. 10.1039/B901735F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Causton HC, Feeney KA, Ziegler CA, et al. : Metabolic Cycles in Yeast Share Features Conserved among Circadian Rhythms. Curr Biol. 2015;25(8):1056–62. 10.1016/j.cub.2015.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Syed S, Saez L, Young MW: Kinetics of doubletime kinase-dependent degradation of the Drosophila period protein. J Biol Chem. 2011;286(31):27654–62. 10.1074/jbc.M111.243618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ruoff P, Loros JJ, Dunlap JC: The relationship between FRQ-protein stability and temperature compensation in the Neurospora circadian clock. Proc Natl Acad Sci U S A. 2005;102(49):17681–6. 10.1073/pnas.0505137102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hirano A, Fu YH, Ptáček LJ: The intricate dance of post-translational modifications in the rhythm of life. Nat Struct Mol Biol. 2016;23(12):1053–60. 10.1038/nsmb.3326 [DOI] [PubMed] [Google Scholar]
- 72. Meng QJ, Maywood ES, Bechtold DA, et al. : Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc Natl Acad Sci U S A. 2010;107(34):15240–5. 10.1073/pnas.1005101107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gallego M, Eide EJ, Woolf MF, et al. : An opposite role for tau in circadian rhythms revealed by mathematical modeling. Proc Natl Acad Sci U S A. 2006;103(28):10618–23. 10.1073/pnas.0604511103 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 74. Shanware NP, Hutchinson JA, Kim SH, et al. : Casein kinase 1-dependent phosphorylation of familial advanced sleep phase syndrome-associated residues controls PERIOD 2 stability. J Biol Chem. 2011;286(14):12766–74. 10.1074/jbc.M111.224014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Vanselow K, Vanselow JT, Westermark PO, et al. : Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes Dev. 2006;20(19):2660–72. 10.1101/gad.397006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lamia KA, Sachdeva UM, DiTacchio L, et al. : AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326(5951):437–40. 10.1126/science.1172156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gao P, Yoo SH, Lee KJ, et al. : Phosphorylation of the cryptochrome 1 C-terminal tail regulates circadian period length. J Biol Chem. 2013;288(49):35277–86. 10.1074/jbc.M113.509604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hirota T, Lee JW, St John PC, et al. : Identification of small molecule activators of cryptochrome. Science. 2012;337(6098):1094–7. 10.1126/science.1223710 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 79. Ode KL, Ukai H, Susaki EA, et al. : Knockout-Rescue Embryonic Stem Cell-Derived Mouse Reveals Circadian-Period Control by Quality and Quantity of CRY1. Mol Cell. 2017;65(1):176–90. 10.1016/j.molcel.2016.11.022 [DOI] [PubMed] [Google Scholar]
- 80. Larrondo LF, Olivares-Yañez C, Baker CL, et al. : Circadian rhythms. Decoupling circadian clock protein turnover from circadian period determination. Science. 2015;347(6221):1257277. 10.1126/science.1257277 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 81. Rosensweig C, Reynolds KA, Gao P, et al. : An evolutionary hotspot defines functional differences between CRYPTOCHROMES. Nat Commun. 2018;9(1):1138. 10.1038/s41467-018-03503-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Patke A, Murphy PJ, Onat OE, et al. : Mutation of the Human Circadian Clock Gene CRY1 in Familial Delayed Sleep Phase Disorder. Cell. 2017;169(2):203–215.e13. 10.1016/j.cell.2017.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 83. Querfurth C, Diernfellner AC, Gin E, et al. : Circadian conformational change of the Neurospora clock protein FREQUENCY triggered by clustered hyperphosphorylation of a basic domain. Mol Cell. 2011;43(5):713–22. 10.1016/j.molcel.2011.06.033 [DOI] [PubMed] [Google Scholar]
- 84. Gustafson CL, Partch CL: Emerging models for the molecular basis of mammalian circadian timing. Biochemistry. 2015;54(2):134–49. 10.1021/bi500731f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gustafson CL, Parsley NC, Asimgil H, et al. : A Slow Conformational Switch in the BMAL1 Transactivation Domain Modulates Circadian Rhythms. Mol Cell. 2017;66(4):447–457.e7. 10.1016/j.molcel.2017.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 86. Aryal RP, Kwak PB, Tamayo AG, et al. : Macromolecular Assemblies of the Mammalian Circadian Clock. Mol Cell. 2017;67(5):770–782.e6. 10.1016/j.molcel.2017.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 87. Ode KL, Ueda HR: Design Principles of Phosphorylation-Dependent Timekeeping in Eukaryotic Circadian Clocks. Cold Spring Harb Perspect Biol. 2018;10(8): pii: a028357. 10.1101/cshperspect.a028357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wong DC, O'Neill JS: Non-transcriptional processes in circadian rhythm generation. Curr Opin Physiol. 2018;5:117–32. 10.1016/j.cophys.2018.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 89. Abe J, Hiyama TB, Mukaiyama A, et al. : Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock. Science. 2015;349(6245):312–6. 10.1126/science.1261040 [DOI] [PubMed] [Google Scholar]
- 90. Tseng R, Goularte NF, Chavan A, et al. : Structural basis of the day-night transition in a bacterial circadian clock. Science. 2017;355(6330):1174–80. 10.1126/science.aag2516 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 91. Mauvoisin D, Wang J, Jouffe C, et al. : Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc Natl Acad Sci U S A. 2014;111(1):167–72. 10.1073/pnas.1314066111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Robles MS, Cox J, Mann M, et al. : In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 2014;10(1):e1004047. 10.1371/journal.pgen.1004047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Narumi R, Shimizu Y, Ukai-Tadenuma M, et al. : Mass spectrometry-based absolute quantification reveals rhythmic variation of mouse circadian clock proteins. Proc Natl Acad Sci U S A. 2016;113(24):E3461–7. 10.1073/pnas.1603799113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Honma S, Yasuda T, Yasui A, et al. : Circadian behavioral rhythms in Cry1/Cry2 double-deficient mice induced by methamphetamine. J Biol Rhythms. 2008;23(1):91–4. 10.1177/0748730407311124 [DOI] [PubMed] [Google Scholar]
- 95. Blum ID, Zhu L, Moquin L, et al. : A highly tunable dopaminergic oscillator generates ultradian rhythms of behavioral arousal. eLife. 2014;3: e05105. 10.7554/eLife.05105 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 96. Bourguignon C, Storch KF: Control of Rest: Activity by a Dopaminergic Ultradian Oscillator and the Circadian Clock. Front Neurol. 2017;8:614. 10.3389/fneur.2017.00614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Susaki EA, Tainaka K, Perrin D, et al. : Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157(3):726–39. 10.1016/j.cell.2014.03.042 [DOI] [PubMed] [Google Scholar]
- 98. Ertürk A, Becker K, Jährling N, et al. : Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc. 2012;7(11):1983–95. 10.1038/nprot.2012.119 [DOI] [PubMed] [Google Scholar]
- 99. Pan C, Cai R, Quacquarelli FP, et al. : Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat Methods. 2016;13(10):859–67. 10.1038/nmeth.3964 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 100. Chung K, Deisseroth K: CLARITY for mapping the nervous system. Nat Methods. 2013;10(6):508–13. 10.1038/nmeth.2481 [DOI] [PubMed] [Google Scholar]
- 101. Yoo SH, Kojima S, Shimomura K, et al. : Period2 3'-UTR and microRNA-24 regulate circadian rhythms by repressing PERIOD2 protein accumulation. Proc Natl Acad Sci U S A. 2017;114(42):E8855–E8864. 10.1073/pnas.1706611114 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 102. Nishide S, Honma S, Honma KI: Two coupled circadian oscillations regulate Bmal1-ELuc and Per2-SLR2 expression in the mouse suprachiasmatic nucleus. Sci Rep. 2018;8(1):14765. 10.1038/s41598-018-32516-w [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation