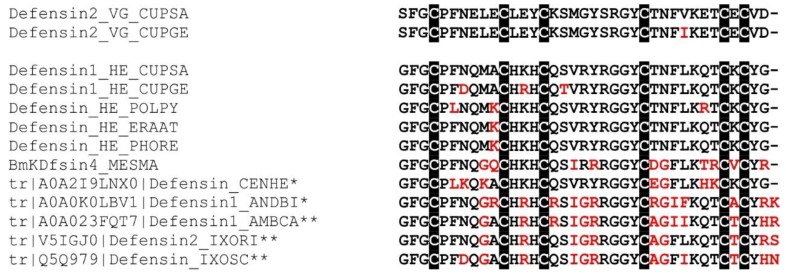
Figure 9.
Multiple sequence alignment of mature defensin-like peptides from different arachnids and types of tissue. Amino acid residue differences of defensins within different tissue subgroups (VG, venom gland; HE, hemocytes; * venom glands, ** salivary glands) are given in red characters and cysteines are highlighted in black. Sequences belong to the spiders C. salei (defensin2_VG_CUPSA, (CSG_98_27980_30560); defensin1_HE_CUPSA [121]), Cupiennius getazi (defensin2_VG_CUPGE, (DN21681_c1_g2_i3_4); defensin1_HE_CUPGE, (DN14943_c0_g1_i1_6)), Polybetes pythagoricus (defensin_HE_POLPY [121]), Eratigena atrica (defensin_HE_ERAAT [121]), Phoneutria reidyi (defensin_HE_PHORE [121]), the scorpions Mesobuthus martensii (BmKDfsin4_MESMA [122]), Centruroides hentzi (tr|A0A2I9LNX0_defensin_CENHE [123]), Androctonus bicolor (tr|A0A0K0LBV1_defensin1_ANDBI [124]), and the ticks Ixodes scapularis (tr|V5IGJ0_defensin2_IXORI [125]), Ixodes ricinus (tr|Q5Q979_defensin_IXOSC), and Amblyomma cajennense (tr|A0A023FQT7_defensin1_AMBCA [120]).