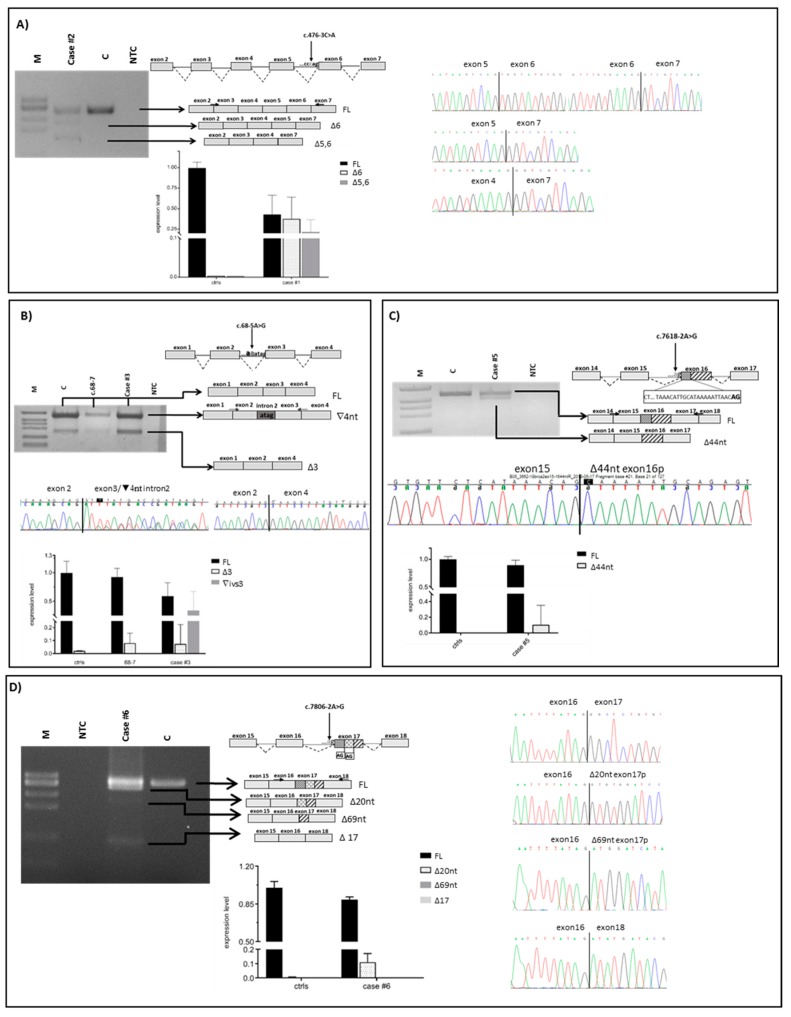
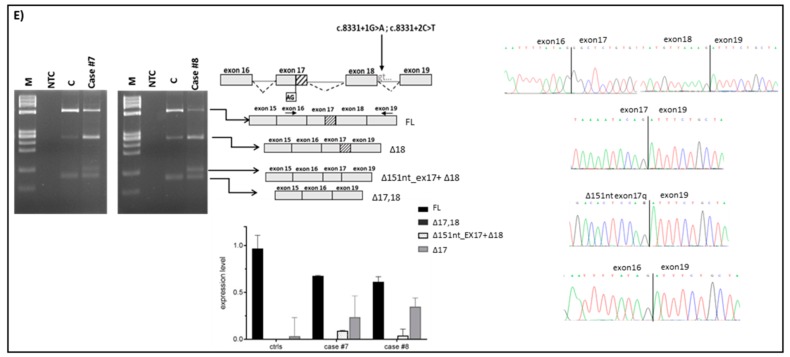
Figure 2.
mRNA analysis of spliceogenic BRCA2 variants. Amplified products were separated on 3% agarose gel and independently sequenced. Gel images, graphical representation, and exon–intron junction sequences of full-length (FL) and aberrant transcripts are shown in upper panels. Variants and RT-PCR primers locations are indicated by the arrows. qPCR results are shown as histograms displaying the relative expression of the full-length and aberrant transcript in the patient’s and controls’ samples (lower panels). (A) c.476-3C>A, causing the up-regulation of the isoforms lacking exon 6 (Δ6) and exons 5 and 6 (∆5,6). The Δ6 transcript accounts for 0.36 (±0.15) and Δ5,6 for 0.23 (±0.15) of overall gene expression in the variant carrier (case #2), and is nearly undetectable in controls. (B) c.68-5A>G variant, causing the retention of four nucleotide at the 3’-end of intron 2 (∇4nt) and the up-regulation of the isoform lacking exon3 (∆3). The aberrant transcript, detectable exclusively in the variant case (case #3), accounts for 0.34 (±0.3) of overall gene expression. The ∆3 isoform accounts for 0.07 (±0.24) of overall gene expression in the variant carrier, 0.08 (±0.08) in the c.68-7T>A sample, and 0.02 (±0.05) in controls. (C) c.7618-2A>G variant, causing the deletion of 44 nt at the 5’-end of exon 16 (∆44nt). The aberrant transcript accounts for 0.10 (±0.24) of overall gene expression in the variant carrier (case #5) and is undetectable in controls. (D) c.7806-2A>G, causing three different aberrant transcripts resulting from the deletion of 20 nt at the 5’end of exon 17 (Δ20nt), of 69 nt at the 5’end of exon 17 (Δ69nt) and of whole exon 17 (Δ17). Only the Δ20nt transcript is detectable in the variant carrier (case #6) and accounts for 0.11 (±0.06) of overall gene expression and is barely detected in control samples. (E) c.8331+1G>A and c.8331+2T>C variants, causing the deletion of 151 nt at the 3’-end of exon 17 and of the whole exon 18 (Δ151nt_ex17 + Δ18), and the up-regulation of the isoforms resulting from the deletion of exon 18 (Δ18) and of exons 17 and 18 (Δ17,18). The ∆151nt_ex17 + ∆18 transcript accounts for 0.09 (±0.001) and 0.04 (±0.06) of overall gene expression in c.8331+1G>A (case #7) and c.8331+2T>C (case #8) samples, respectively, and is undetectable in controls. The ∆18 transcript accounts for 0.23 (±0.2), 0.34 (± 0.09) and 0.03 (±0.21) of overall gene expression in case #7, case #8 and controls, respectively. The ∆17,18 transcript is undetectable in both variant cases and controls. M, molecular weight marker (0.15–2.1 Kbp, Roche and ΦX174 DNA/BsuRI (HaeIII) Marker, Thermo Scientific); C, healthy control sample; NTC, no template control.