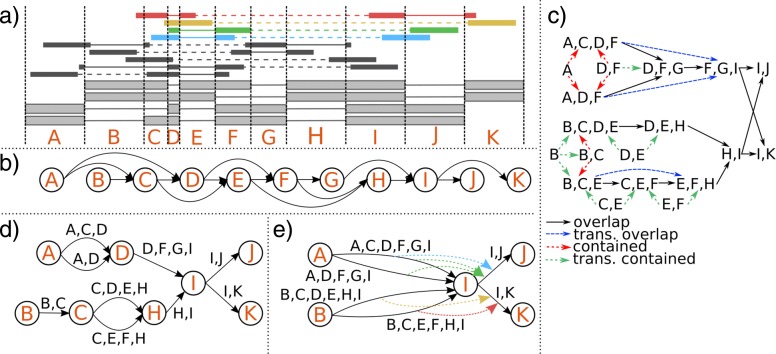
Fig. 1.
Overview of the most important steps of the transcript assembly pipeline. In (a) an example set of transcripts over the same gene is shown, together with potential paired-end reads leading to them. Reads have the same length r, but varying insert lengths. Be |X| the number of basepairs in exon X, then: |A|,|C|,|D|,|E|,|F|,|G|<r and |C|+|D|<r. All other conditions are >r. The commonly used split graph (b) removes all evidence from reads spanning more than two exons and paired-end information. Instead, an exon-bin based subset — adding edges between bins when a bin is a subset of another bin — and overlap — adding edges when a bin’s suffix is prefix of other bin — graph (c) is created (trivial subsets omitted). After removing transitive components, a directed acyclic splice graph is created that allows each bin to be uniquely mapped to a set of edges, but minimizes the number of ambiguous connections (d). Any maximum- or minimum-flow implementation can be used to establish transcript expressions on each edge. Due to flow conservation, composite paths and tree nodes can be reduced without loss of information. Flow decomposition into final transcripts can be improved by using evidences (e) from multi-exon-spanning single reads or paired-end data (dashed edge-links, color-code by (a))