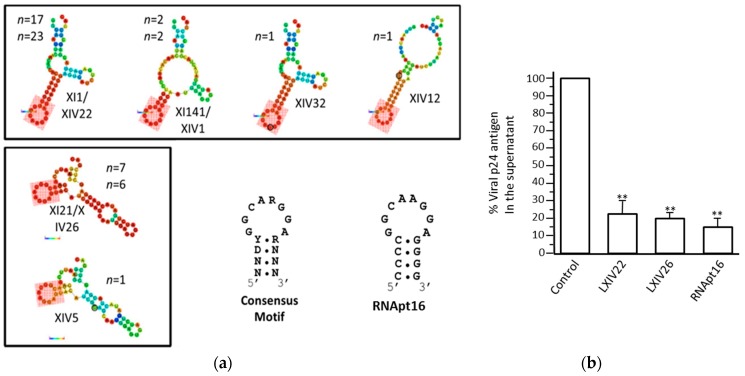
Figure 8.
The in silico-designed RNApt16: (a) The structural analysis of the representative examples of 64-nt-long selected aptamers shows the minimum free energy isoform. The probabilities of every nucleotide to actually hold the structural conformation shown are represented by a colour code (red, highest; dark blue, lowest). The pink, shadowed box indicates the consensus 16-nt-long structural domain. The sequence and secondary structure of the consensus motif and the designed RNApt16 are indicated at the bottom. R, purine; Y, pirimidine; N, any ribonucleotide, (b) HIV-1 inhibition assays: The inhibition of HIV-1 particles production measured as the p24 antigen production by the XIV22, XIV26, and RNApt16 aptamers (compared to a negative control). The data represent the mean of three independent assays. ** The significant differences as compared to the control (p < 0.01). The figure is adapted from Reference [69].