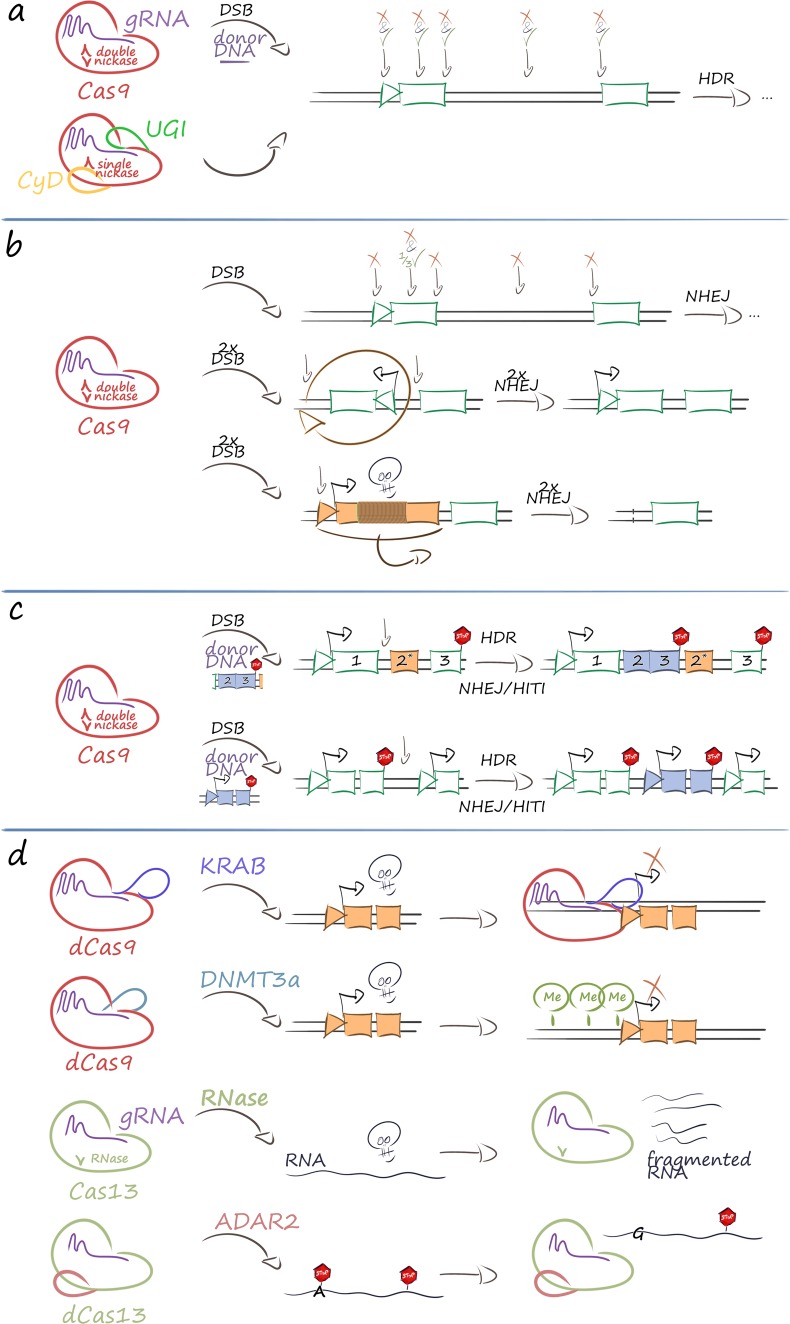
Fig. 1.
Therapeutic strategies based on CRISPR/Cas-based tools. The therapeutic use of CRISPR/Cas ribonucleoprotein complexes is wide-ranging and in its present application falls into four main categories. a Mutation-specific precision editing of any suitable nucleotide can be performed either by the original DNA endonuclease (double nickase) Cas9 molecule and HDR-mediated repair or by DNA base editors. This can be used to correct disease-causing mutations (green tick) or introduce disabling sequence changes (red cross). The figure illustrates as exemplary positions (from left to right) changes in the promoter region, the coding sequence, the splice donor site, deep intronic regulatory sequences, and the splice acceptor site. b Sequence disruption and rearrangement can be achieved by double nickases and NHEJ-based mechanisms. Top: sequences can be disrupted by a single DSB in the promoter region, the coding sequence, the splice donor site, deep intronic regulatory sequences, and the splice acceptor site. For some applications the statistical one-third of in-frame insertion and/or deletion (indel) events for changes in the open reading frame may be corrective and exploited for therapy. Middle: pathogenic inversions in the genome can be reversed by two flanking DSBs and NHEJ-mediated re-ligation (shown). Alternatively, inversion or excision can disable pathogenic or unwanted regulatory regions (not shown). Bottom: deletions for the removal of pathogenic or unwanted regulatory regions can likewise be achieved by two flanking DSBs and NHEJ-mediated re-ligation (shown), but has also been demonstrated by an appropriately spaced pair of single nickases (not shown). c Targeted integration in the genome can be achieved by a single DSB and, depending on the precision required at the junctions, can employ HDR-mediated repair or NHEJ-mediated HITI. Top: insertion of a cDNA under control of an endogenous promoter is one application of targeted integration; integration in intronic sequences of the mutated gene will allow repair of mutations downstream from the DSB. Bottom: insertion of expression cassettes into safe harbor loci, such as an intergenic region (shown) or the intron of an endogene (not shown) that will allow stable transgene expression, can be used for safe gene addition. d Modulation of gene expression can be performed at multiple levels and may employ, from top to bottom, deactivated Cas9 fused to transcriptional regulators (e.g., in order to dynamically repress expression of pathogenic genes), deactivated Cas9 fused to epigenome modifiers (e.g., in order to permanently repress expression of pathogenic genes by DNA methylation), Cas13-like RNA-guided RNA nucleases (e.g., in order to dynamically knock down mRNA of pathogenic genes), and nuclease-deficient Cas13-like fusions to chemical RNA-base modifiers (e.g., in order to repair nonsense mutations by A>G conversion). For clarity, epigenetic modifiers for DNA demethylation, histone acetylation, histone deacetylation, and others are not shown. Likewise, RNA base conversion for disease-causing missense mutations (such as demonstrated Ala>Trp conversion) is not shown. Throughout, the skull indicates a pathogenic gene or molecule and the STOP sign indicates a translation termination (nonsense) codon. ADAR2 adenosine deaminase acting on RNA 2 [82], cDNA complementary DNA, CRISPR/Cas clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas), CyD cytidine deaminase domain for C>U conversion in the single-stranded DNA loopout, currently with precision of ≤ 2 base pairs, dCas9 deactivated Cas9 without endonuclease activity, DNMT3a catalytic domain of DNA methyltransferase 3 alpha for DNA methylation and potentially persistent repression of gene expression for affected promoters [78], DSB double-strand break, gRNA guide RNA, HDR homology-directed repair, HITI homology-independent targeted integration, indel insertion and/or deletion, KRAB catalytic domain of Krüppel-associated box epigenetic repressor [80], mRNA messenger RNA, NHEJ non-homologous end-joining, UGI uracil DNA glycosylase inhibitor domain to prevent base excision repair and removal of base edit