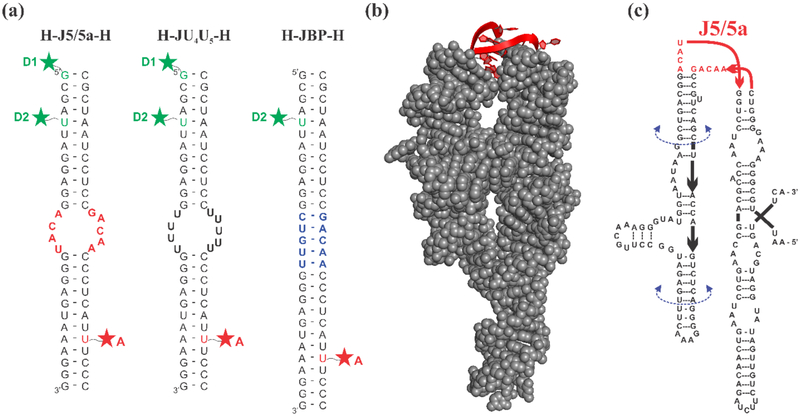
Figure 1:
(a) Schematic of three RNA constructs. Constructs consist of two RNA oligonucleotides which are individually labeled with a fluorophore and annealed together. The resulting construct forms either two short duplexes connected by an asymmetric internal loop, or a fully base paired control. Green and red stars represent the dye label positions, which are achieved from either 5’ C12-amino modifier (D1) or an amino-modified dT replacing the uracil nucleotide in the sequence (D2 and A). The constructs are named according to the nature of the junction at the center. H-J5/5a-H: the junction has the native J5/5a sequence found in P4-P6. H-JU4U5-H: both junction strands are replaced by uracil nucleotides. H-JBP-H: the junction contains 5 Watson Crick base pairs. (b) The crystal structure of the full-length wild-type P4-P6 RNA (PDB: 1GID) with the J5/5a junction shown in red. (c) The sequence of the wild-type P4-P6 RNA. The U-mutant P4-P6 replaces the J5/5a junction sequence with 9 uracil nucleotides. The base-paired P4-P6 replaces “ACAU” with “CUGUU” forming 5 canonical base pairs within the junction region.