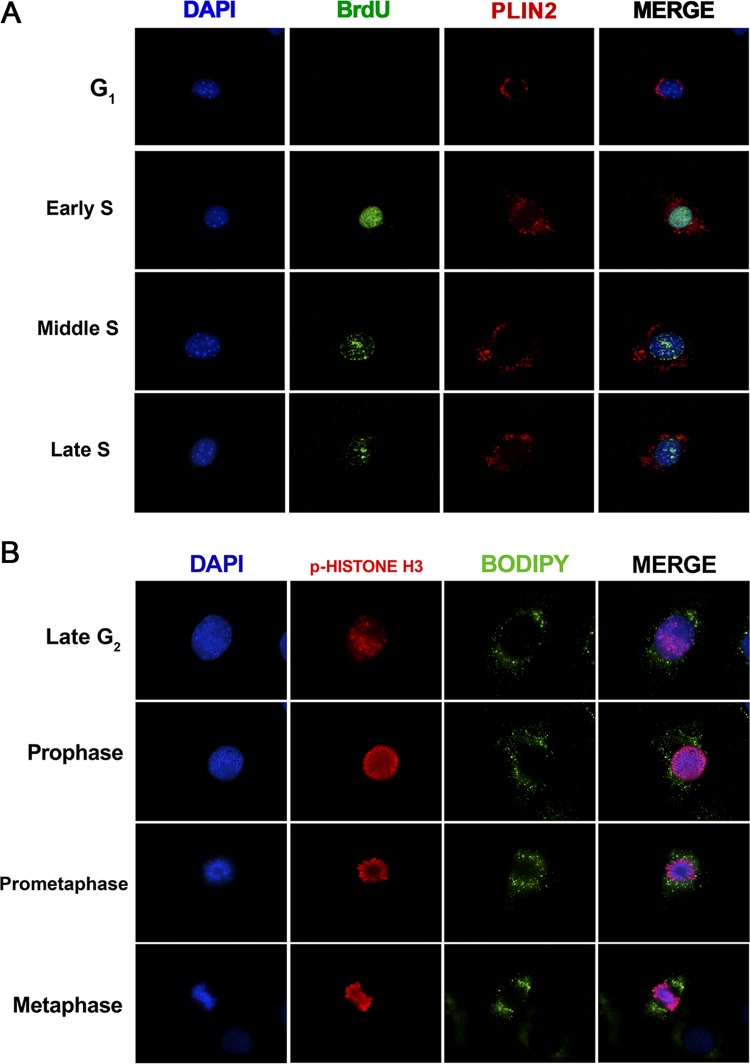
FIG 4.
The subcellular localization of lipid droplets is highly dynamic during different cell cycle phases. NIH 3T3 cells were synchronized by confluence and serum starvation as shown in Fig. 1A and were then replated at a low density and supplemented with a medium containing 10% FBS. (A) Synchronized NIH 3T3 cells on coverslips were incubated with 20 μM BrdU for 30 min before fixation at the indicated time points. Later, cells were treated with anti-BrdU for the detection of DNA synthesis (green) and with anti-PLIN2 for lipid body staining (red), along with DAPI for nuclear staining (blue). Fluorescence microscopy analysis shows the subcellular localization of lipid droplets and BrdU nuclear staining at the indicated time points. (B) Synchronized NIH 3T3 cells were fixed on coverslips at 48 h after serum supplementation and were then treated with antibodies to phospho-histone H3 (p-histone H3) for the detection of chromosome condensation (red) and with Bodipy for lipid droplet staining (green), along with DAPI for nuclear staining (blue). Fluorescence microscopy analysis shows the subcellular localization of lipid droplets during G2 and mitosis. All data are representative of the results of at least three independent experiments.