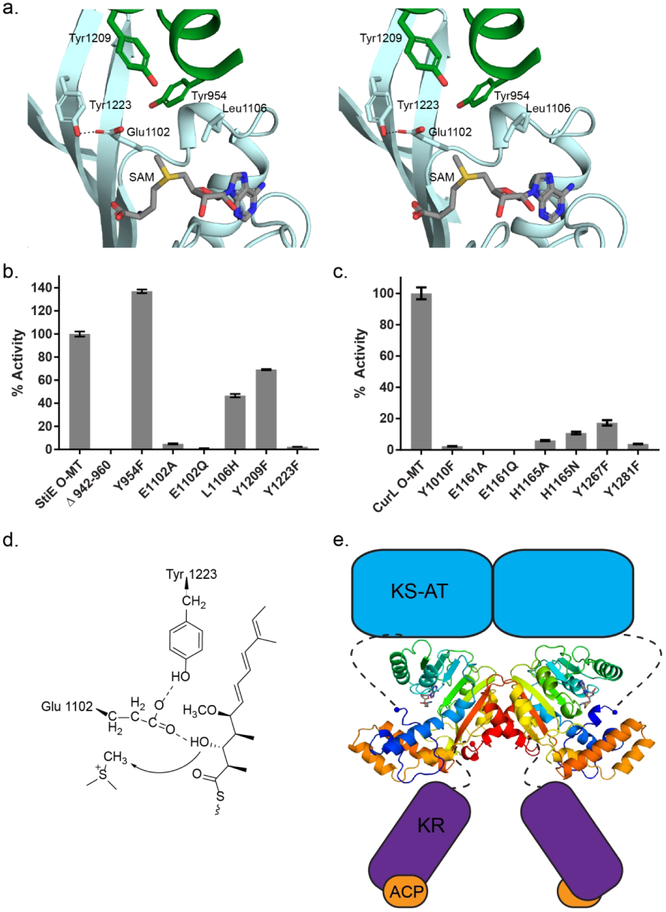
Figure 4.
Relative methylation activities of CurL and StiE O-MT variants. (a) StiE O-MT active site colored as in Fig. 3d. Substituted amino acids are shown in sticks. (b) StiE O-MT activity on (3R)-3-hydroxy-5-methoxy-myristoyl-ACP substrate (6). (c) CurL O-MT activity on (R)-3 hydroxydodecanoyl-ACP substrate (8). Activity relative to wild-type was quantified using the LC-MS based Ppant ejection assay.34, 35 The mutagenesis sites in StiE and CurL O-MTs are analogous (StiE/CurL: Tyr954/Tyr1010, Glu1102/Glu1161, Leu1106/His1165, Tyr1209/Tyr1267, Tyr1223/Tyr1281) based on alignment of the 35% identical sequences. Error bars represent triplicate experiments. (d) StiE O-MT mechanism. The conserved Glu polarizes the β-hydroxyl for methylation. (e) Model of StiD module. Connections of the O-MT to neighboring domains are indicated with dashed lines. For the dimeric StiD and StiE O-MTs, the linker from AT is 30–35 amino acids and linker to KR is 14–18 amino acids.