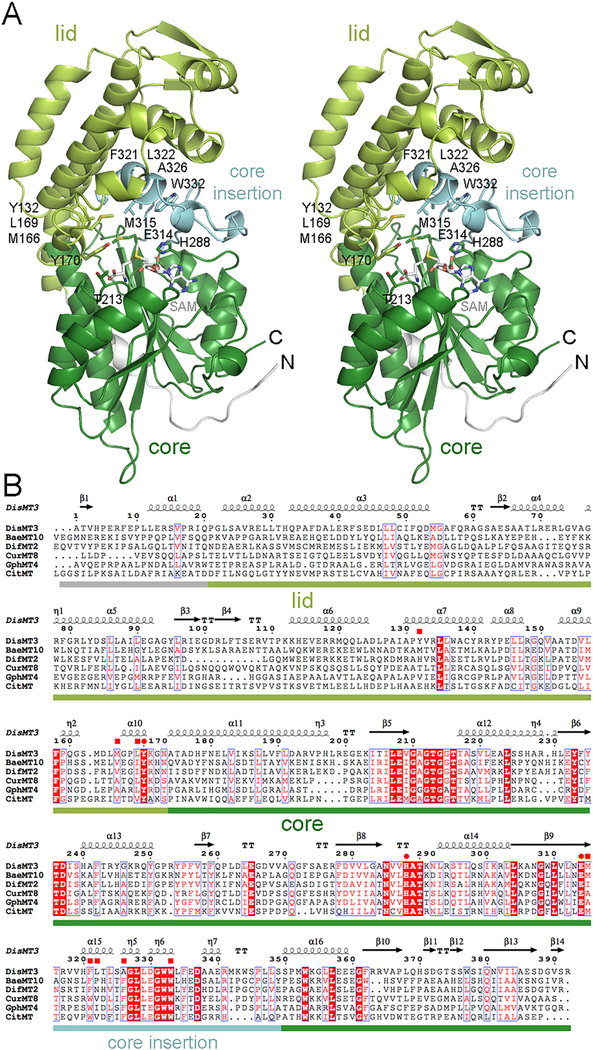
Figure 2.
Structure of an MT from a trans-AT assembly line. (A) A stereodiagram shows the architecture of DisMT3. The 22 N-terminal residues, the lid subdomain, the core subdomain, and the core insertion are indicated by color. SAM was modeled into the active site based on the location of SAH in CitMT (PDB 5MPT). Residues thought to position the substrate for catalysis are shown as sticks. (B) A sequence alignment shows the similarity of MTs from trans-AT assembly lines (DisMT3, BaeMT10, and DifMT2), cis-AT assembly lines (CurMT8 and GphMT4), and iterative PKSs (CitMT). The invariant glutamate/histidine dyad as well as a highly conserved tyrosine are indicated with circles, while residues that help form the substrate tunnel are indicated with squares. DisMT3, disorazol, Sorangium cellulosum So ce12, CAI43932; BaeMT10, bacillaene, Bacillus amyloliquefaciens FZB42, CAG23959; DifMT2, difficidin, Bacillus amyloliquefaciens FZB42, CAG23977; CurMT8, curacin, Moorea producens 3L, AEE88280; GphMT4, gephronic acid, Archangium violaceum Cb vi76, AHA38200; CitMT, citrinin, Monascus purpureus, BAD44749.