FIG 1.
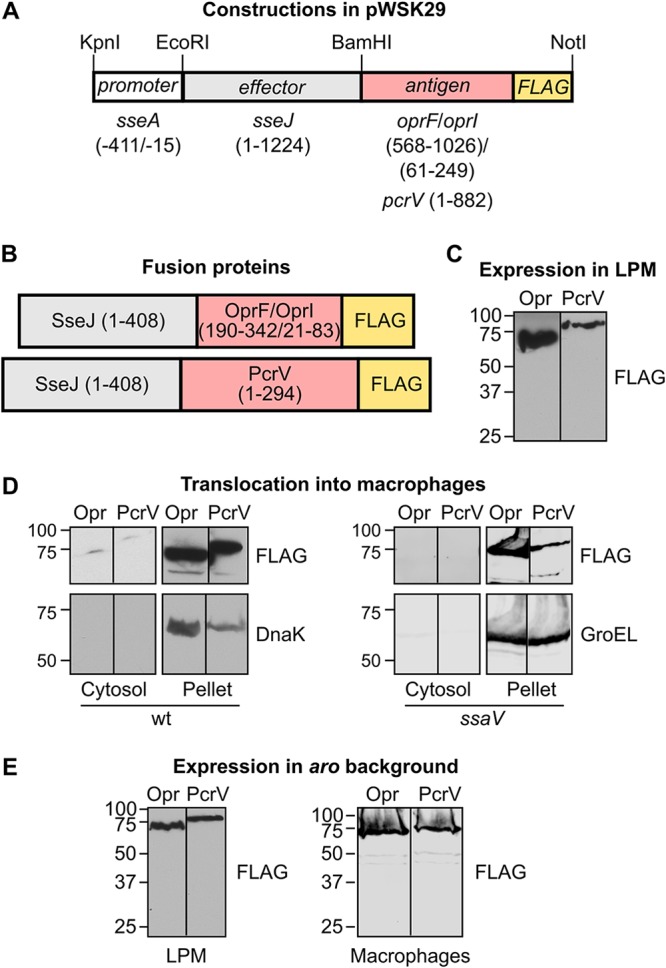
Generation of protein fusions for delivery of Pseudomonas aeruginosa antigens. (A) Vector pWSK29 was used for the generation of two plasmids in three steps. In the first step, part of the coding region of antigens OprF/OprI (see Materials and Methods for the generation of the hybrid gene oprF-oprI) or the complete coding region (without the stop codon) for the antigen PcrV from P. aeruginosa was amplified by PCR. DNA encoding the FLAG epitope and recognition sites for BamHI and NotI endonucleases was added with the primers used for amplification. These amplicons were cleaved with BamHI and NotI and ligated into pWSK29 previously cleaved with the same restriction enzymes. In the second step, the coding region of sseJ without the stop codon was amplified by PCR and ligated to the plasmids obtained in the first step previously cleaved with EcoRI and BamHI restriction enzymes. Finally, the promoter region of sseA (nucleotides −411 to −15 relative to the translation start site) was amplified by PCR and added to the plasmids obtained in the second step previously cleaved with endonucleases KpnI and EcoRI. Numbers in parentheses indicate base pairs relative to the start of the coding region. (B) Fusion proteins that are expected to be produced under the appropriate conditions after the introduction of the plasmids described in panel A in S. Typhimurium. Numbers in parentheses refer to amino acids included in the fusions. Both fusions include the complete amino acid sequence of SseJ and a C-terminal FLAG epitope to facilitate identification of the fusions by Western blotting. The expected molecular masses are 70.18 kDa for SseJ-OprF/I-FLAG and 79.23 kDa for SseJ-PcrV-FLAG. (C) Derivatives of S. Typhimurium strain 14028 carrying plasmids encoding OprF/I (Opr) or PcrV fusion proteins were grown in LPM. A FLAG tag was used for detection of the proteins by immunoblotting. Molecular weights in kDa are indicated on the left. (D) Salmonella strains that were derivatives of the wild-type strain (wt) or the ssaV mutant (carrying a nonfunctional T3SS2) expressing SseJ in fusion with Pseudomonas antigens OprF/I (Opr) or PcrV with a FLAG tag were grown under noninvasive conditions. These bacteria were used to infect RAW264.7 cells for 8 h. Cells were lysed with 1% Triton X-100 in PBS and centrifuged 15 min at 15,000 × g. The pellets and the filtered concentrated supernatants (cytosol) were analyzed by immunoblotting with anti-FLAG antibodies to detect translocation of fusion proteins into the host cytosols. Incubations with anti-DnaK or anti-GroEL antibodies were used as controls of contamination of the cytosol fraction with nonsecreted bacterial proteins. (E) Expression of the fusion proteins SseJ-OprF/I (Opr) and SseJ-PcrV (PcrV) was analyzed in an aroA aroB (aro) background. Bacteria were grown in LPM (left) or were grown for 24 h at 37°C with shaking in LB medium and used to infect RAW264.7 macrophages (right). Protein extracts were analyzed by immunoblotting with an anti-FLAG antibody.
