FIGURE 2.
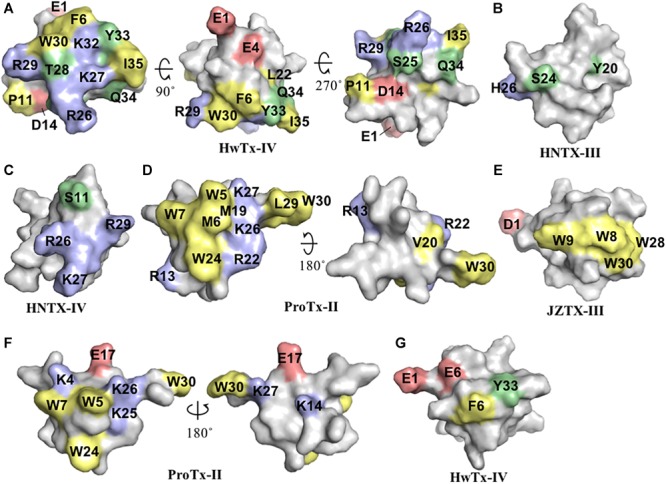
Three-dimensional structure of spider ICK peptides displaying key residues involved in the inhibition of NaV channels and cell membrane binding. (A) Three-dimensional structure of HwTx-IV determined by NMR (PBD 2m4x) (Minassian et al., 2013), (B) HNTX-III determined by NMR (PBD 2jtb), (C) HNTX-IV determined by NMR (PBD 1niy) (Li et al., 2004), (D) ProTx-II determined by X-Ray (PBD 5o0u) (Wright et al., 2017), and (E) JZTX-III determined by NMR (PBD 2i1t). The labeled amino acids residues have key role in potency over NaV channels and lead to loss in activity as described in the text and Table 1. (F) Structure of ProTx-II determined by X-Ray (PDB 5o0u) (Wright et al., 2017) and (G) Structure of HwTx-IV (PDB 2m4x) (Minassian et al., 2013) determined by NMR. The labeled amino acids residues have key role in cell membrane binding as described in the text and Table 1. Amino acids residues are colored as follow: yellow for hydrophobic, red for acid, blue for basic and green for neutral. All three-dimensional structures were prepared in PyMOL (DeLano, 2002).
