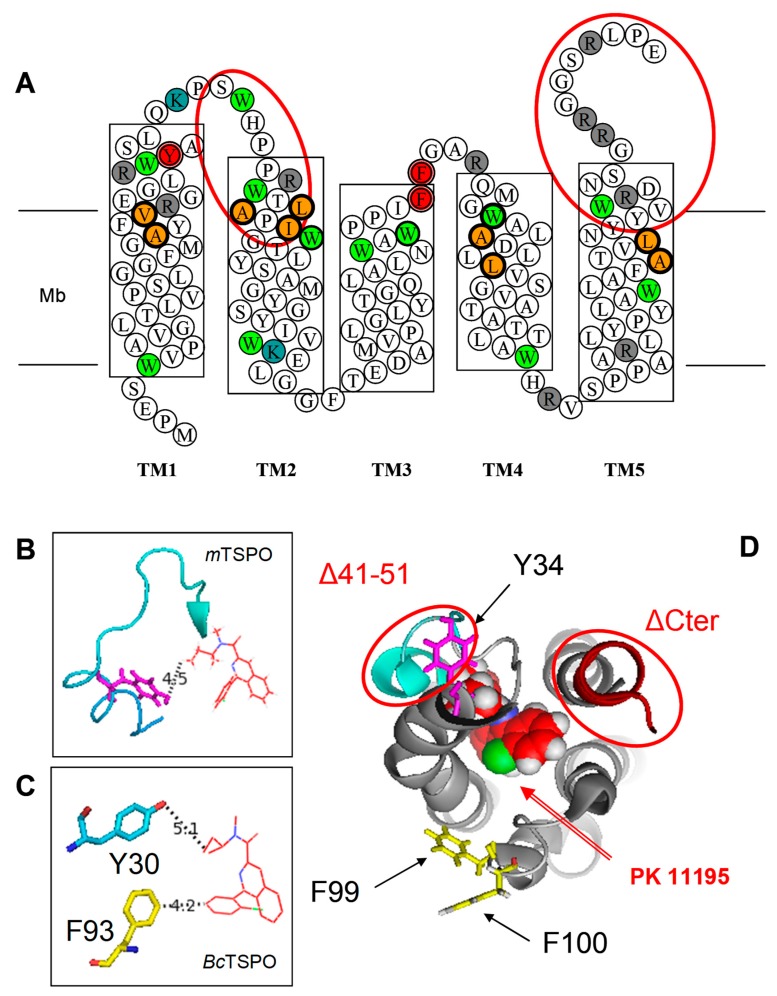
Figure 3.
(A) The mTSPO sequence in a 2-D diagram with transmembrane helices shown as boxes crossing the membrane (Mb): The point mutations and deletion mutants used in the present work are shown as red-coloured double circles or ellipses, respectively. The amino acids involved in the binding pocket of the atomic structure are shown in orange-filled bold circles. Lysine (K), arginine (R) and tryptophan (W) are shown in blue-, grey- and green-filled circles, respectively. (B) The position of Y34 in the NMR structure of A147T polymorph of mammalian TSPO [18] (PDB ID-2NO2) and (C) the position of Y30 and F93 in the X-ray structure of bacterial TSPO [16] (PDB ID-4RYI): These two residues are homologous to positions Y32 and F100 of mammalian TSPO. (D) The top view of atomic NMR structure with bound PK 11195 [9] (PDB ID-2MGY) emphasizes the positions of the mutations and deletions.