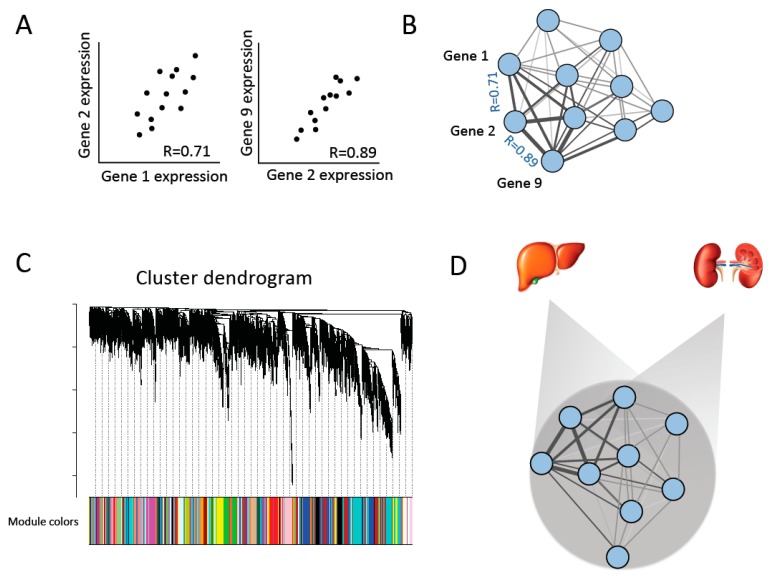
Figure 1.
Constructing gene co-expression networks [29]. (A) Co-expression similarity is compared between pairs of genes among individuals in order to build (B) a co-expression network of all genes. (C) Co-expression modules are identified and defined by hierarchical clustering and cutting branches off the dendrogram. Modules are then assigned colors for identification. (D) Consensus networks across each pair of tissues are created to identify co-expression modules that are conserved across tissues (consensus modules) [31].