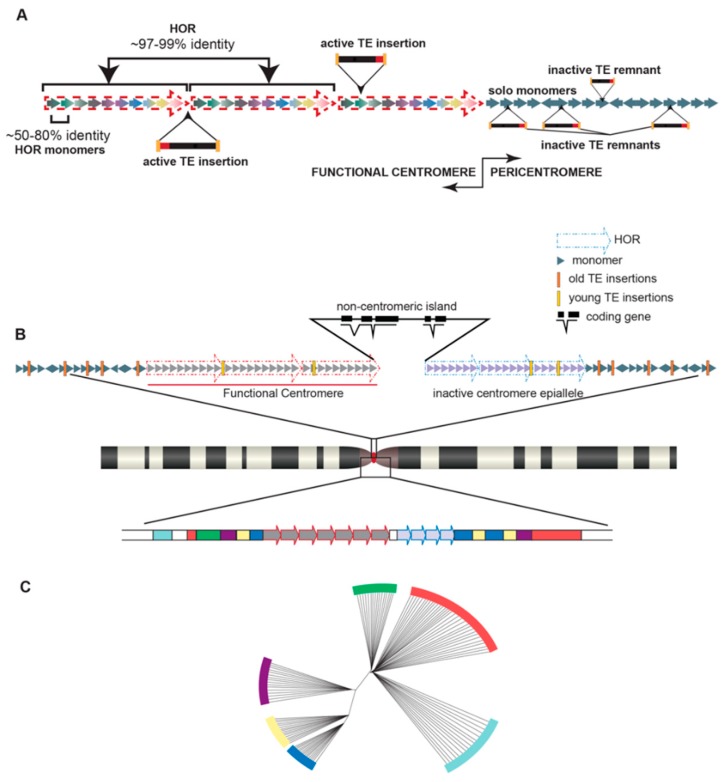
Figure 1.
Overview of satellite DNA structure in a human centromere/pericentromere. (a) α satellite monomers (colored solid arrows) are organized into a repeating unit, called a higher order repeat (HOR) (red dashed arrows). In this example, 10 monomers are in each HOR (10-mers). HOR units are repeated in a chromosome-specific manner 100–1000 s of times within a functional centromere core. Within a single HOR, monomers share anywhere from 50–80% sequence identity with one another. The same monomer within different HORs in the same array may share up to 99% identity. Solo monomers (solid arrows) are found in the pericentromeric region and are highly variable in terms of sequence and orientation. Within the centromere, transposable elements (TE) insertions typically include recently active or active (hot) elements, while the TE insertions found in the pericentromere are older, inactive elements. (b) The core centromere structure (red dot, chromosome schematic) of human chromosomes (a generic chromosome ideogram is indicated, middle) consists of different α satellite arrays arranged in HORs (dashed arrows). Each HOR array may contain a different monomer number; in this example, the functional centromere (i.e., assembles CENP-A nucleosomes) at a 10-mer HOR (red dashed arrows). A 7-mer HOR is found nearby but is an inactive epiallele. Both HORs are separated by non-centromeric DNA, which may contain genes. α satellites are also found throughout the pericentromere (bottom schematic, different colored blocks). (c) Representative cladogram of the phylogenetic relationship of the non-HOR α monomers shown in (b). In this example, strata of newer satellites are closer to the HOR arrays, while older satellites are found more distally. Relative age of satellites is indicated by tree branch length; shorter branches are younger elements and deeper branches are older.