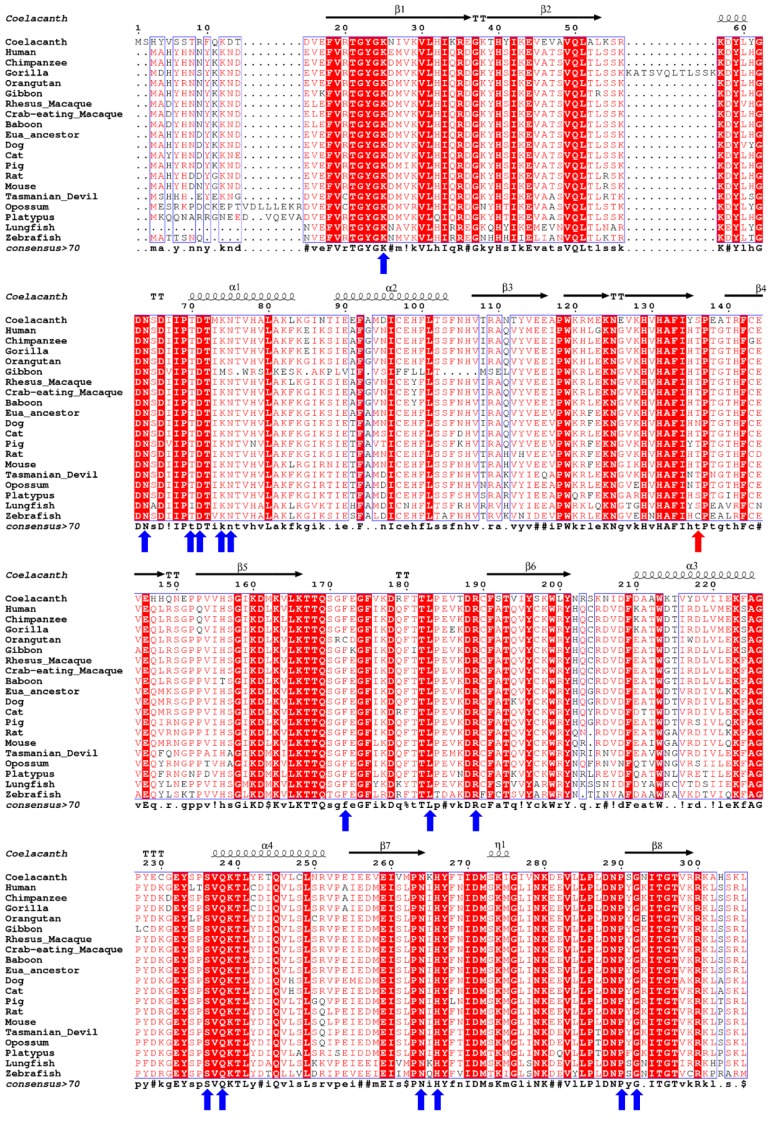
Figure 1.
Multiple sequence alignment of coelacanth uricase (UOX) with UOX from related organisms. The secondary structure shown on top of the sequences is predicted by ESpript 3.0 [29] using PDB ID: 4MB8 as a template. Helices and strands are named according to Retailleau et al [28]. Consensus amino acids are shown under the sequences. Residues related to the active sites are indicated with blue arrows. Zebrafish C129, which forms natural disulphide bridges is indicated with a red arrow. Cat UOX sequence is started at 27th amino acid. The human sequence is deduced by replacing the two stop points (caused by the two premature stop codons 33 and 187) with arginine.