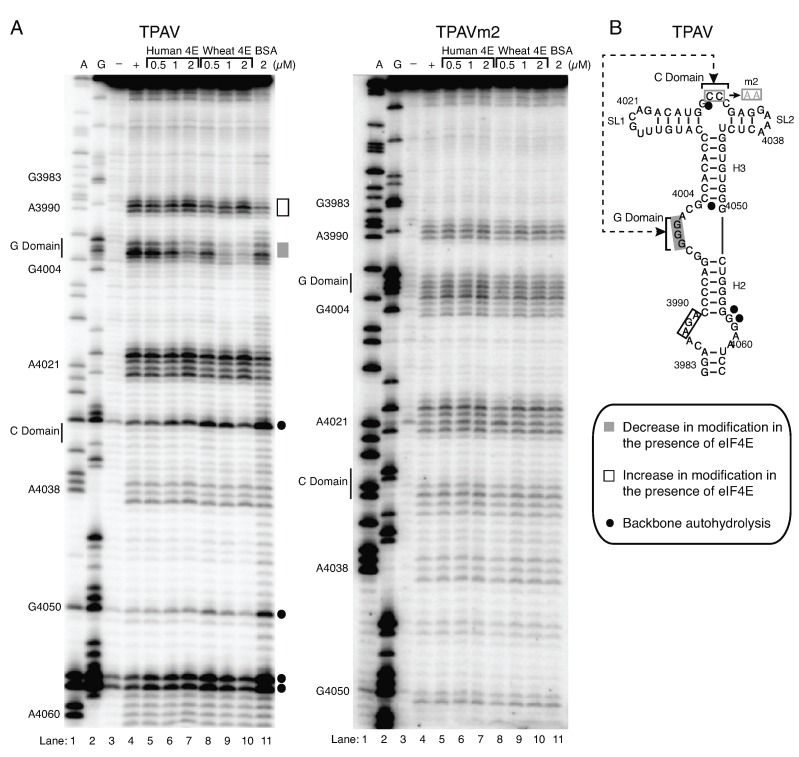
Figure 4.
(A) TPAV and TPAVm2 PTE modification and cleavage structure probing patterns generated using the SHAPE reagent benzoyl cyanide (BzCN) in the absence of protein (+ lane) or presence of indicated proteins (lanes 5–11). A and G are dideoxy sequencing lanes, with positions of selected bases indicated at left. Unmodified RNA (− lane) shows background RNA hydrolysis. Gray and white bars to the right show regions with increased or decreased, respectively, modification by BzCN in the presence of eIF4E. Filled circles at right denote regions of autohydrolysis in the absence of BzCN. (B) Positions of human eIF4E-induced changes in SHAPE sensitivity mapped onto the TPAV PTE secondary structure. Double-headed arrow indicates predicted pseudoknot base pairing between the C and G domains, which is disrupted by the CC to AA changes in mutant m2 (boxed in gray).