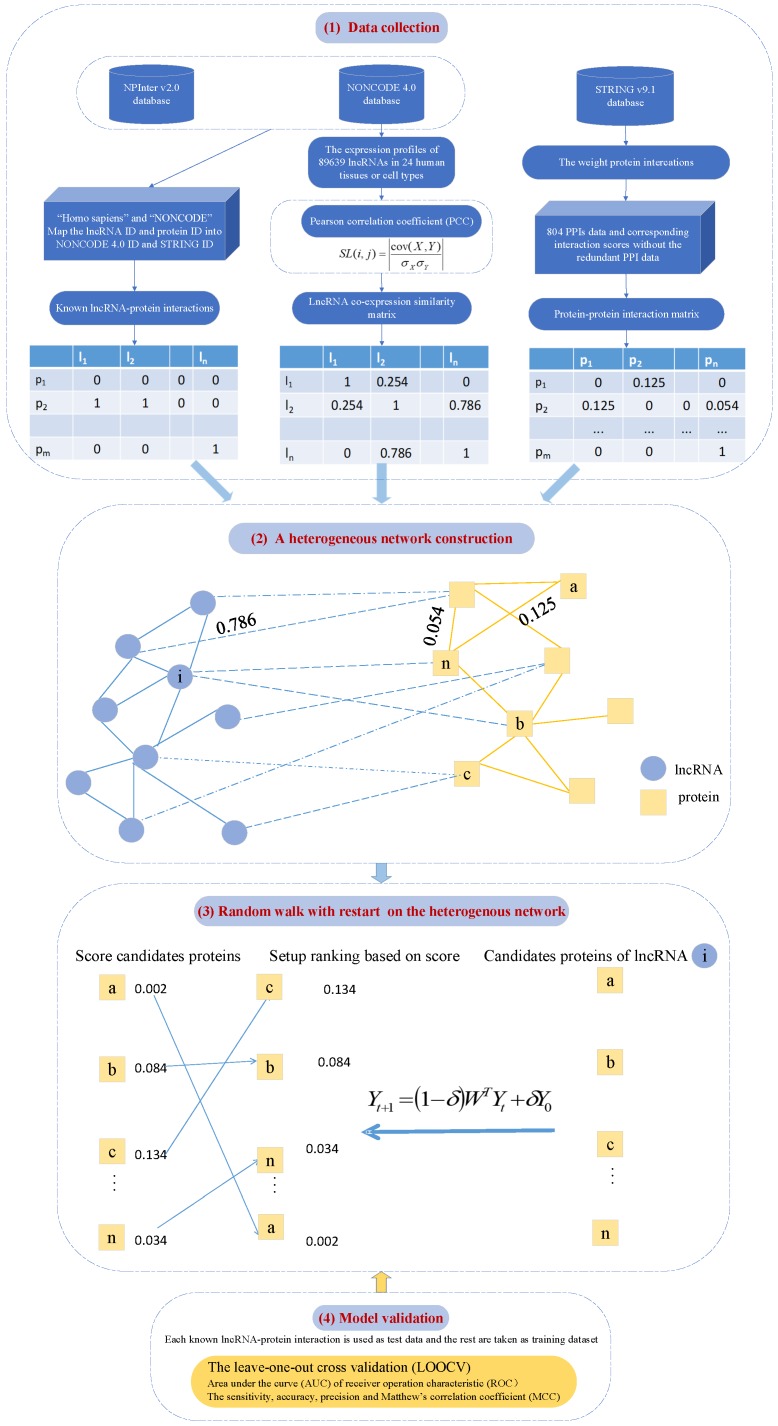
Figure 5.
Pipeline of LPIHN, containing three modules: (1) Data collection: lncRNA–protein interactions from NPInter, protein–protein interactions from STRING database and lncRNA–lncRNA similarity network computed based on lncRNA expression profile from NONCODE. (2) A heterogeneous network construction. (3) LncRNA–protein interactions prediction based on the random walk with restart. A score is assigned to each candidate protein of a query lncRNA, by the random walk with restart on the heterogeneous network. The candidate proteins are ranked based on the scores. (4) Model validations based on LOOCV and AUC. For LPIHN, the lncRNA–lncRNA similarity network is calculated by using the lncRNA expression profiles based on the PCC of each pair of lncRNAs. The heterogeneous network is constructed by connecting the lncRNA–lncRNA similarity network and PPI network together with the known lncRNA–protein interaction network. Blue circles indicated lncRNAs, orange squares indicated proteins, blue edges indicated lncRNA–lncRNA similarities, orange edges indicated protein–protein interactions, and blue dotted edges indicated known lncRNA–protein interactions.