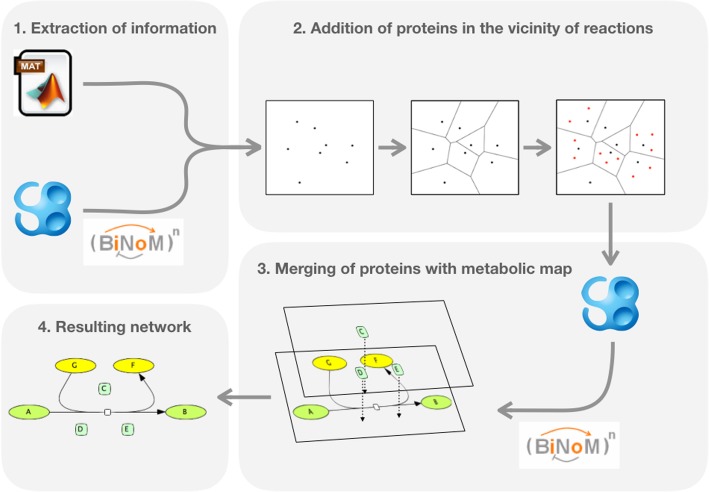
Fig. 1.
General workflow for integration of proteins into a metabolic network. (1) Extraction of the informations on proteins present in metabolic reactions from a model and CellDesigner file. (2) Addition of proteins in the vicinity of catalysed reactions. (3) Merging of obtained proteins with the metabolic map through the BiNoM plugin. (4) As a result, a CellDesigner network file containing proteins on top of the original metabolic network is obtained. This file can be later integrated into NaviCell through the NaviCell Factory tool