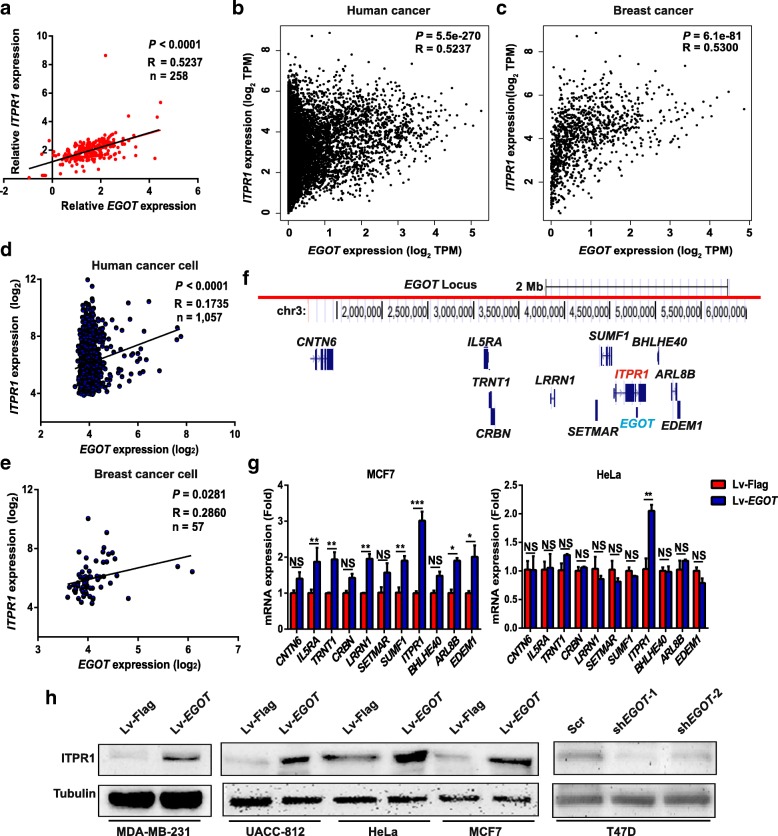
Fig. 2.
EGOT expression is positively related to ITPR1 expression. a Correlation between EGOT and ITPR1 expression in 258 breast cancer tissues in the HMUCC cohort. EGOT and ITPR1 levels (normalized to GAPDH) were subjected to Pearson’s correlation analysis. b, c Correlation between EGOT and ITPR1 expression in 33 cancer types (n = 9664) (b) and in breast cancer (n = 1085) (c). Data were downloaded from the TCGA portal, and the expression levels of EGOT and ITPR1 are indicated by TPM values (log2). d, e Correlation between EGOT and ITPR1 expression in 1057 human cancer cell lines (d) and 57 breast cancer cell lines (e). Data were downloaded from the CCLE. EGOT and ITPR1 levels (normalized to GAPDH) were subjected to Pearson’s correlation analysis. f Schematic illustration of genes near the EGOT locus (less than 2 Mb). Location information was obtained from the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway). g Expression of nearby genes in MCF7 EGOT overexpression cells (left) and HeLa EGOT overexpression cells (right). Data are shown as the means ± s.d. Student’s t-test was applied for statistical analysis: * P < 0.05; ** P < 0.01; and *** P < 0.001. h Protein expression levels of ITPR1 in EGOT overexpression and knockdown cells. Data represent at least three independent experiments