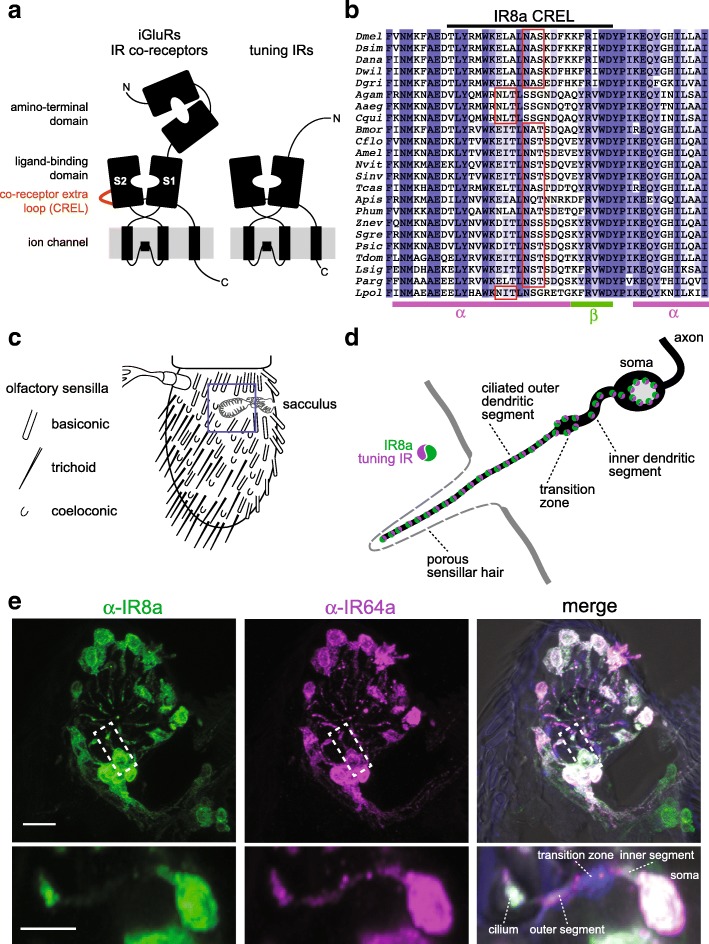
Fig. 1.
The IR co-receptors contain a distinctive N-glycosylated loop. a Schematic of the domain organisation of iGluRs, IR co-receptors and tuning IRs. b Alignment of the protein sequence spanning the CREL (co-receptor extra loop; black bar) of IR8a orthologues from the indicated species. Predicted N-glycosylation sites are highlighted with red boxes and predicted secondary structure is shown below the alignment. Species (top-to-bottom): Drosophila melanogaster, Drosophila simulans, Drosophila ananassae, Drosophila willistoni, Drosophila grimshawi, Anopheles gambiae, Aedes aegypti, Culex quinquefasciatus, Bombyx mori, Camponotus floridanus, Apis mellifera, Nasonia vitripennis, Solenopsis invicta, Tribolium castaneum, Acyrthosiphon pisum, Pediculus humanus, Zootermopsis nevadensis, Schistocerca gregaria, Phyllium siccifolium, Thermobia domestica, Lepismachilis y-signata, Panulirus argus, Limulus polyphemus. c Schematic of the Drosophila third antennal segment showing the distribution of different olfactory sensilla and the internal sacculus. d Schematic illustrating the main anatomical features of an olfactory sensory neuron (OSN); the morphology of the cuticular hair and the branched nature of the cilium varies between different sensilla classes (note: most sensilla contain more than one neuron per hair). e Immunofluorescence with antibodies against IR8a (green) and IR64a (magenta) on an antennal section of a wild-type animal, showing the region containing the third chamber of the sacculus (blue boxed area in c). In the merged image, the transition zone is marked by monoclonal antibody 21A6 (blue), and a bright-field image is overlaid to reveal cuticular anatomical landmarks. Scale bar: 10 μm. The images shown below are of a single OSN (from a subset of optical slices of the area indicated by the dashed white boxes) in which the main anatomical features are shown. Scale bar: 5 μm