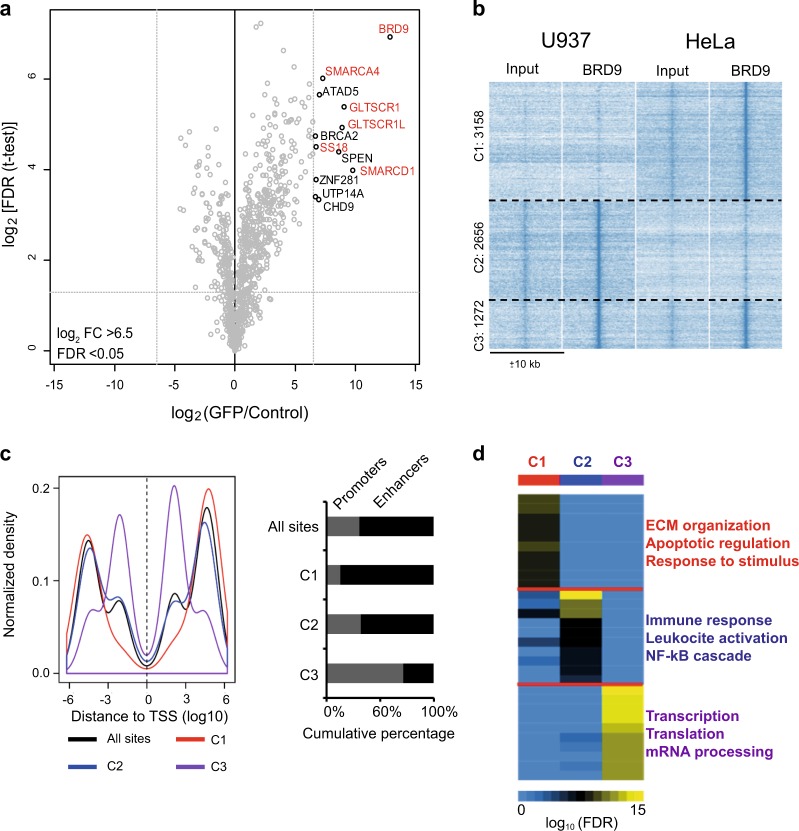
Fig. 4. Genome-wide binding of BRD9 is highly cell type specific.
a Volcano plot from label-free GFP pulldown of GFP-BRD9 HeLa cell nuclear extracts. Bait and its interactors are shown in the upper right corner (SWI/SNF members including BRD9 are shown in red). Statistically, enriched proteins in GFP-BRD9 pulldown were identified by a permutation-based FDR-corrected t-test. Label-free quantification intensity of GFP pull-down relative to control (fold change, x-axis) is plotted against the log2-transformed p-value of t-test (y-axis). BRD9 interacts with proteins previously reported as core member of the BAF-SWI/SNF complex. b K-means clustering analysis of BRD9 ChIP-seq data showing three distinct clusters of BRD9-binding sites with different specificity for U937 and HeLa cells. c Distance of the clusterized BRD9-binding sites to TSS (promoters: ≤1 kb from TSS, putative enhancers: >1 kb from TSS). d GO analysis of BRD9-binding sites using Genome regions enrichment of annotations tool (GREAT). The most significant GO terms for each of the three clusters are shown