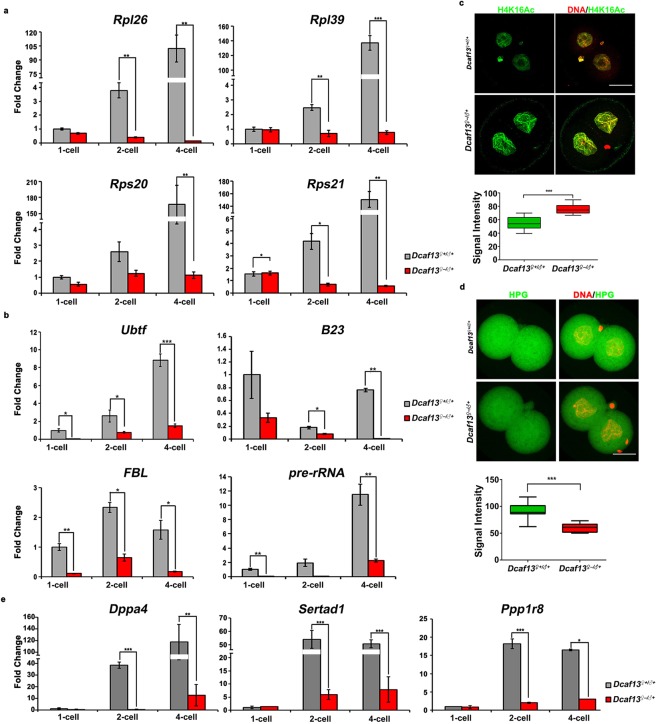
Figure 4.
Absence of maternal DCAF13 abrogates zygotic nucleus function in transcription and results in decreased translation. (a,b) Graphical representation of the mean expression level ± standard error of the mean (SEM) by reverse transcription polymerase chain reaction (RT-PCR) for several ribosomal protein genes (a) and rDNA processing factor gene (b) in Dcaf13♀+/♂+ (in gray) or Dcaf13♀−/♂+ (in red) zygotes, two-cell and four-cell (two-cell stage in Dcaf13♀−/♂+) embryos. *p < 0.05, ** to p < 0.01 and*** to p < 0.001. (c) Immunofluorescence using antibodies against H4K16Ac (in green) and DNA (in red) in two-cell embryos (top), and intensity measurement (bottom). Scale bar, 20 µm. *** to p < 0.001. Dcaf13♀+/♂+ (n = 16), Dcaf13♀−/♂+ (n = 16). (d) Detection of newly synthesized protein by homopropargylglycine (HPG) incorporation in two-cell embryos of Dcaf13♀+/♂+ or Dcaf13♀−/♂+ was shown (top), also with the intensity measurement (bottom). Scale bar, 20 µm. *** to p < 0.001. Dcaf13♀+/♂+ (n = 14), Dcaf13♀−/♂+ (n = 14). (e) Graphical representation of the mean expression level ± SEM by RT-PCR for several representative zygotic genes in Dcaf13♀+/♂+ (in gray) or Dcaf13♀−/♂+ (in red) zygotes, two-cell and four-cell (two-cell stage in Dcaf13♀−/♂+) embryos. *p < 0.05, ** to p < 0.01, and *** to p < 0.001.